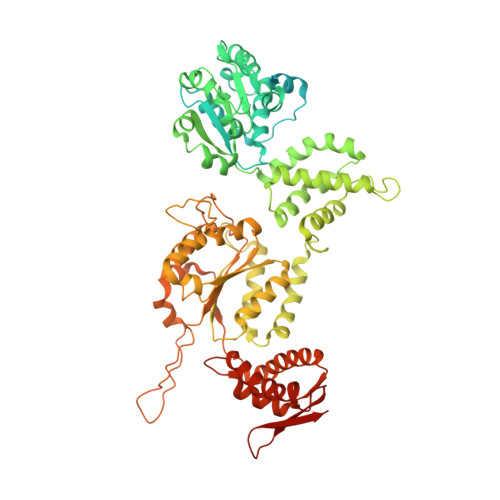
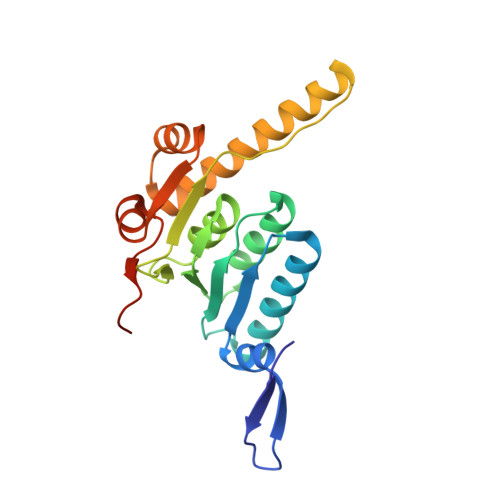
AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Kim, S., Fei, X., Sauer, R.T., Baker, T.A.(2022) Nat Struct Mol Biol 29: 1068-1079
- PubMed: 36329286
- DOI: https://doi.org/10.1038/s41594-022-00850-3
- Primary Citation of Related Structures:
7UIV, 7UIW, 7UIX, 7UIY, 7UIZ, 7UJ0 - PubMed Abstract:
ClpAP, a two-ring AAA+ protease, degrades N-end-rule proteins bound by the ClpS adaptor. Here we present high-resolution cryo-EM structures of Escherichia coli ClpAPS complexes, showing how ClpA pore loops interact with the ClpS N-terminal extension (NTE), which is normally intrinsically disordered. In two classes, the NTE is bound by a spiral of pore-1 and pore-2 loops in a manner similar to substrate-polypeptide binding by many AAA+ unfoldases. Kinetic studies reveal that pore-2 loops of the ClpA D1 ring catalyze the protein remodeling required for substrate delivery by ClpS. In a third class, D2 pore-1 loops are rotated, tucked away from the channel and do not bind the NTE, demonstrating asymmetry in engagement by the D1 and D2 rings. These studies show additional structures and functions for key AAA+ elements. Pore-loop tucking may be used broadly by AAA+ unfoldases, for example, during enzyme pausing/unloading.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Organizational Affiliation: