Activation of the human insulin receptor by non-insulin-related peptides
Kirk, N.S., Chen, Q., Wu, Y.G., Asante, A.L., Hu, H., Espinosa, J.F., Martinez-Olid, F., Margetts, M.B., Mohammed, F.A., Kiselyov, V.V., Barrett, D.G., Lawrence, M.C.(2022) Nat Commun 13: 5695
- PubMed: 36171189
- DOI: https://doi.org/10.1038/s41467-022-33315-8
- Primary Citation of Related Structures:
7U6D, 7U6E, 8DI2 - PubMed Abstract:
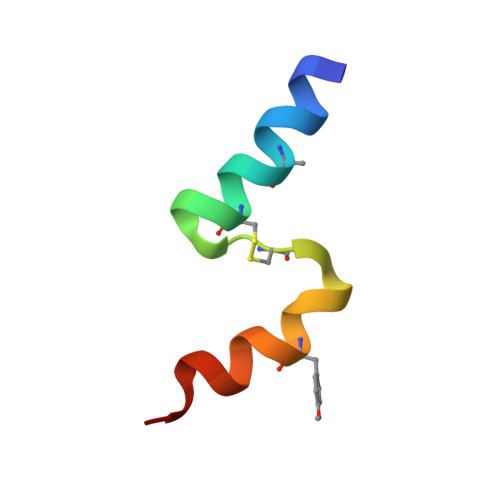
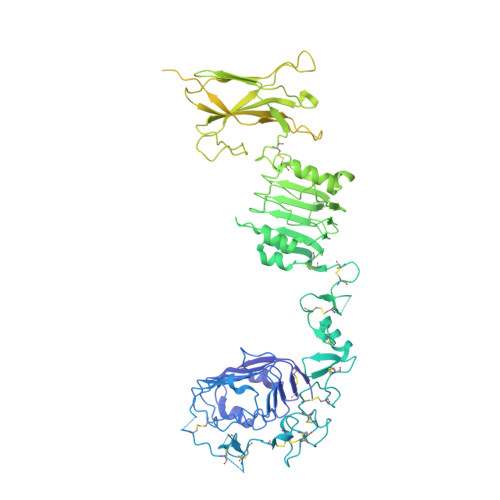
The human insulin receptor signalling system plays a critical role in glucose homeostasis. Insulin binding brings about extensive conformational change in the receptor extracellular region that in turn effects trans-activation of the intracellular tyrosine kinase domains and downstream signalling. Of particular therapeutic interest is whether insulin receptor signalling can be replicated by molecules other than insulin. Here, we present single-particle cryoEM structures that show how a 33-mer polypeptide unrelated to insulin can cross-link two sites on the receptor surface and direct the receptor into a signalling-active conformation. The 33-mer polypeptide engages the receptor by two helical binding motifs that are each potentially mimicable by small molecules. The resultant conformation of the receptor is distinct from-but related to-those in extant three-dimensional structures of the insulin-complexed receptor. Our findings thus illuminate unexplored pathways for controlling the signalling of the insulin receptor as well as opportunities for development of insulin mimetics.
- WEHI, 1G Royal Parade, Parkville, VIC, 3052, Australia.
Organizational Affiliation: