High-valency Anti-CD99 Antibodies Toward the Treatment of T Cell Acute Lymphoblastic Leukemia.
Romero, L.A., Hattori, T., Ali, M.A.E., Ketavarapu, G., Koide, A., Park, C.Y., Koide, S.(2021) J Mol Biology 434: 167402-167402
- PubMed: 34958778
- DOI: https://doi.org/10.1016/j.jmb.2021.167402
- Primary Citation of Related Structures:
7SFX - PubMed Abstract:
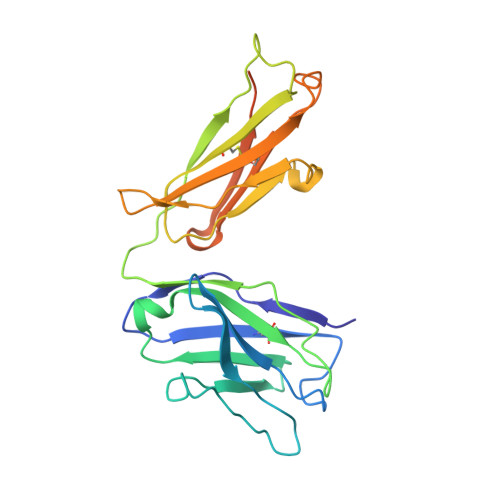
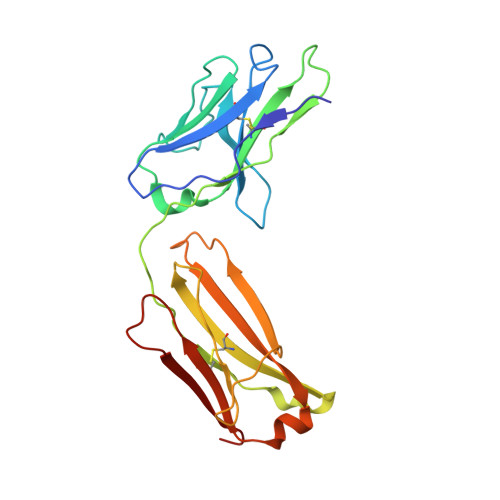
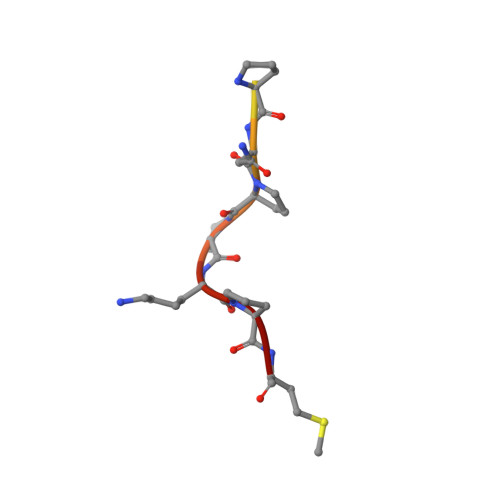
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive form of leukemia that currently requires intensive chemotherapy. While childhood T-ALL is associated with high cure rates, adult T-ALL is not, and both are associated with significant short- and long-term morbidities. Thus, less toxic and effective strategies to treat T-ALL are needed. CD99 is overexpressed on T-ALL blasts at diagnosis and at relapse. Although targeting CD99 with cytotoxic antibodies has been proposed, the molecular features required for their activity are undefined. We identified human antibodies that selectively bound to the extracellular domain of human CD99, and the most potent clone, 10A1, shared an epitope with a previously described cytotoxic IgM antibody. We engineered clone 10A1 in bivalent, trivalent, tetravalent, and dodecavalent formats. Increasing the antibody valency beyond two had no effects on binding to T-ALL cells. In contrast, a valency of ≥3 was required for cytotoxicity, suggesting a mechanism of action in which an antibody clusters ≥3 CD99 molecules to induce cytotoxicity. We developed a human IgG-based tetravalent version of 10A1 that exhibited cytotoxic activity to T-ALL cells but not to healthy peripheral blood cells. The crystal structure of the 10A1 Fab in complex with a CD99 fragment revealed that the antibody primarily recognizes a proline-rich motif (PRM) of CD99 in a manner reminiscent of SH3-PRM interactions. This work further validates CD99 as a promising therapeutic target in T-ALL and defines a pathway toward the development of a selective therapy against T-ALL.
- Department of Biochemistry and Molecular Pharmacology, New York University Grossman School of Medicine, New York, NY 10016, USA.
Organizational Affiliation: