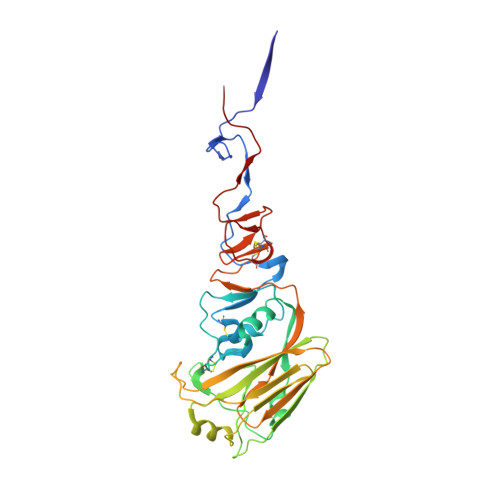
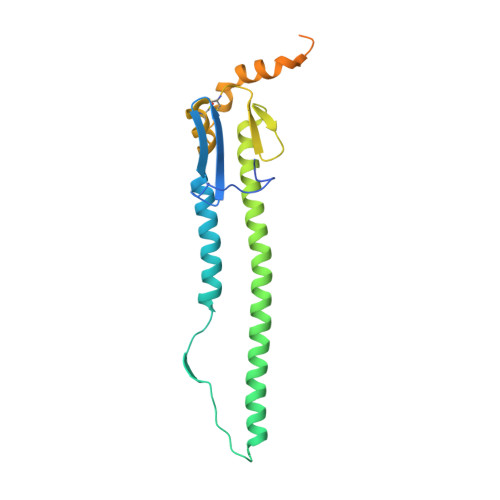
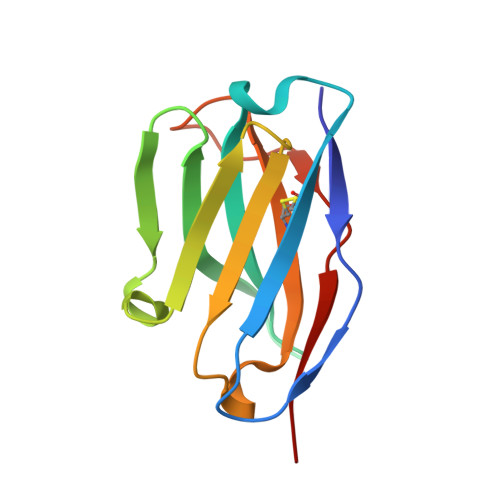
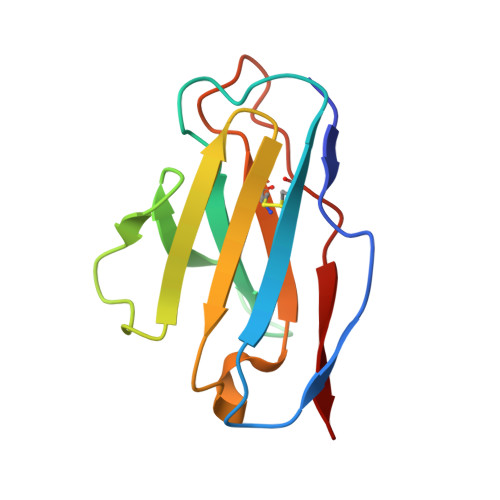
Allelic polymorphism controls autoreactivity and vaccine elicitation of human broadly neutralizing antibodies against influenza virus.
Sangesland, M., Torrents de la Pena, A., Boyoglu-Barnum, S., Ronsard, L., Mohamed, F.A.N., Moreno, T.B., Barnes, R.M., Rohrer, D., Lonberg, N., Ghebremichael, M., Kanekiyo, M., Ward, A., Lingwood, D.(2022) Immunity 55: 1693-1709.e8
- PubMed: 35952670
- DOI: https://doi.org/10.1016/j.immuni.2022.07.006
- Primary Citation of Related Structures:
7SCN, 7SCO - PubMed Abstract:
Human broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin stalk of group 1 influenza A viruses (IAVs) are biased for IGHV1-69 alleles that use phenylalanine (F54) but not leucine (L54) within their CDRH2 loops. Despite this, we demonstrated that both alleles encode for human IAV bnAbs that employ structurally convergent modes of contact to the same epitope. To resolve differences in lineage expandability, we compared F54 versus L54 as substrate within humanized mice, where antibodies develop with human-like CDRH3 diversity but are restricted to single V H genes. While both alleles encoded for bnAb precursors, only F54 IGHV1-69 supported elicitation of heterosubtypic serum bnAbs following immunization with a stalk-only nanoparticle vaccine. L54 IGHV1-69 was unproductive, co-encoding for anergic B cells and autoreactive stalk antibodies that were cleared from B cell memory. Moreover, human stalk antibodies also demonstrated L54-dependent autoreactivity. Therefore, IGHV1-69 polymorphism, which is skewed ethnically, gates tolerance and vaccine expandability of influenza bnAbs.
- The Ragon Institute of Massachusetts General Hospital, The Massachusetts Institute of Technology and Harvard University, 400 Technology Square, Cambridge, MA 02139, USA.
Organizational Affiliation: