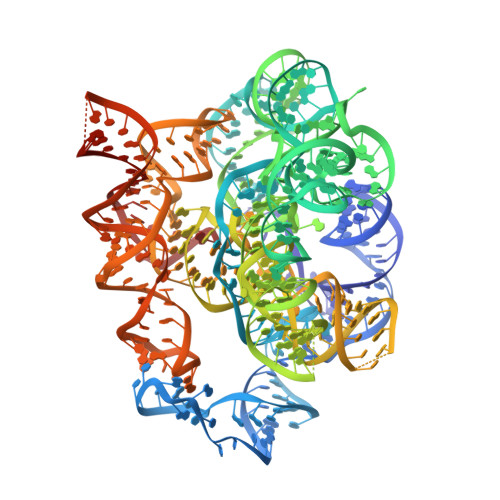
Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Liu, D., Thelot, F.A., Piccirilli, J.A., Liao, M., Yin, P.(2022) Nat Methods 19: 576-585
- PubMed: 35501384
- DOI: https://doi.org/10.1038/s41592-022-01455-w
- Primary Citation of Related Structures:
7R6L, 7R6M, 7R6N - PubMed Abstract:
High-resolution structural studies are essential for understanding the folding and function of diverse RNAs. Herein, we present a nanoarchitectural engineering strategy for efficient structural determination of RNA-only structures using single-particle cryogenic electron microscopy (cryo-EM). This strategy-ROCK (RNA oligomerization-enabled cryo-EM via installing kissing loops)-involves installing kissing-loop sequences onto the functionally nonessential stems of RNAs for homomeric self-assembly into closed rings with multiplied molecular weights and mitigated structural flexibility. ROCK enables cryo-EM reconstruction of the Tetrahymena group I intron at 2.98-Å resolution overall (2.85 Å for the core), allowing de novo model building of the complete RNA, including the previously unknown peripheral domains. ROCK is further applied to two smaller RNAs-the Azoarcus group I intron and the FMN riboswitch, revealing the conformational change of the former and the bound ligand in the latter. ROCK holds promise to greatly facilitate the use of cryo-EM in RNA structural studies.
- Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, USA.
Organizational Affiliation: