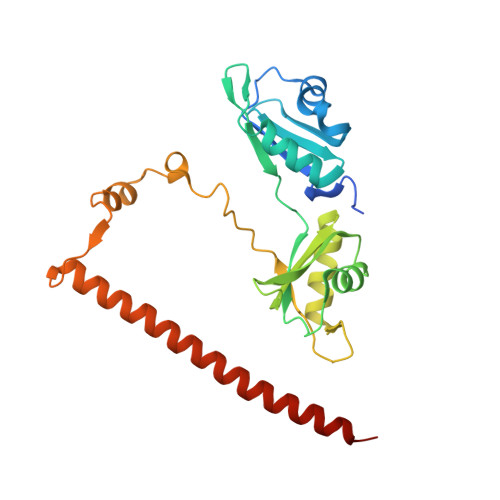
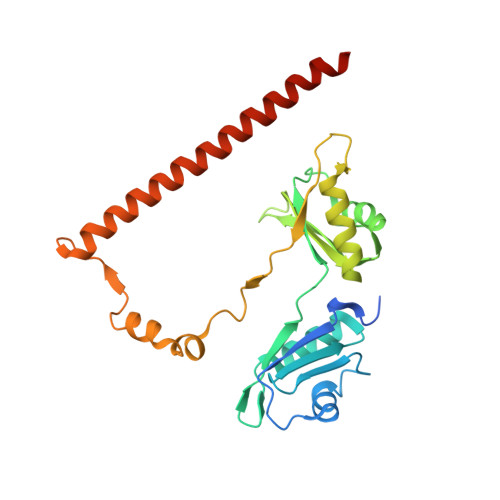
Crystal structure of SFPQ-NONO heterodimer.
Schell, B., Legrand, P., Fribourg, S.(2022) Biochimie 198: 1-7
- PubMed: 35245601
- DOI: https://doi.org/10.1016/j.biochi.2022.02.011
- Primary Citation of Related Structures:
7PU5 - PubMed Abstract:
The Drosophila behavior/human splicing (DBHS) protein family is composed of the three members SFPQ, NONO and PSPC1. These proteins share a strong sequence and structural homology within the core-structured domains forming obligate homo- and heterodimers. This feature may lead to the simultaneous existence of six different dimeric complexes that sustain their function in many cellular processes such as pre-mRNA splicing, innate immunity, transcriptional regulation. In order to perform a complete structural analysis of all possible DBHS dimers, we have solved the crystal structure of the missing DBHS heterodimer SFPQ-NONO at 3.0 Å resolution. We identify subtle changes in amino acid composition and local secondary structure of the NOPS region orientation that may modulate affinity between complexes. Interestingly this area is found mutated in aggressive skin cancers and adenocarcinomas.
- INSERM U1212 - CNRS 5320 & Université de Bordeaux, 146 Rue Léo Saignat, 33000, Bordeaux, France; Universität Konstanz, 78457, Konstanz, Germany.
Organizational Affiliation: