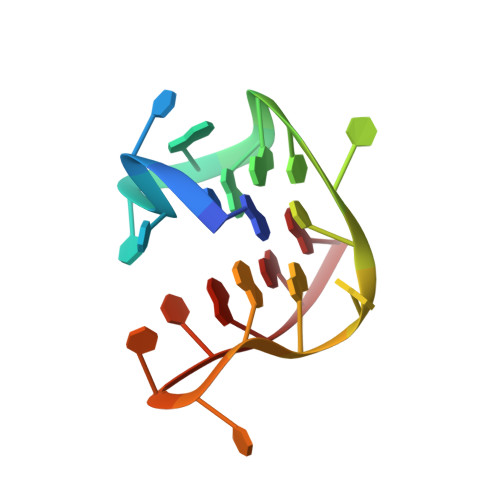
G-quadruplex structure of the C. elegans telomeric repeat: a two tetrads basket type conformation stabilized by a non-canonical C-T base-pair.
Marquevielle, J., De Rache, A., Vialet, B., Morvan, E., Mergny, J.L., Amrane, S.(2022) Nucleic Acids Res 50: 7134-7146
- PubMed: 35736226
- DOI: https://doi.org/10.1093/nar/gkac523
- Primary Citation of Related Structures:
7OQT - PubMed Abstract:
The Caenorhabditis elegans model has greatly contributed to the understanding of the role of G-quadruplexes in genomic instability. The GGCTTA repeats of the C. elegans telomeres resemble the GGGTTA repeats of the human telomeres. However, the comparison of telomeric sequences (Homo sapiens, Tetrahymena, Oxytricha, Bombyx mori and Giardia) revealed that small changes in these repeats can drastically change the topology of the folded G-quadruplex. In the present work we determined the structure adopted by the C. elegans telomeric sequence d[GG(CTTAGG)3]. The investigated C. elegans telomeric sequence is shown to fold into an intramolecular two G-tetrads basket type G-quadruplex structure that includes a C-T base pair in the diagonal loop. This work sheds light on the telomeric structure of the widely used C. elegans animal model.
- Univ. Bordeaux, Inserm U1212, CNRS UMR 5320, ARNA laboratory, 146 rue Léo Saignat F-33000 Bordeaux, France.
Organizational Affiliation: