E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP
Ahel, J., Fletcher, A.J., Grabarczyk, D., Roitinger, E., Deszcz, L., Lehner, A., Virdee, S., Clausen, T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| E3 ubiquitin-protein ligase RNF213 | 4,836 | Mus musculus | Mutation(s): 0 Gene Names: Rnf213, Mystr EC: 2.3.2.27 (PDB Primary Data), 3.6.4 (PDB Primary Data), 2.3.2 (UniProt) | 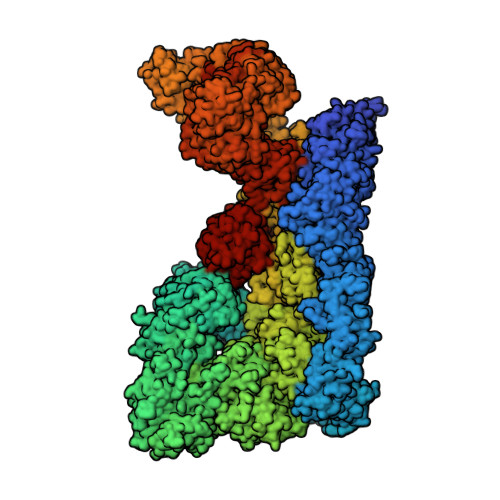 | |
UniProt | |||||
Find proteins for E9Q555 (Mus musculus) Explore E9Q555 Go to UniProtKB: E9Q555 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E9Q555 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS (Subject of Investigation/LOI) Query on AGS | G [auth A] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| ATP (Subject of Investigation/LOI) Query on ATP | B [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | F [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | D [auth A], E [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3.1 |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council (ERC) | Austria | 694978 |
| Austrian Research Promotion Agency | Austria | 852936 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_UU_12016/8 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/ P003982/1 |