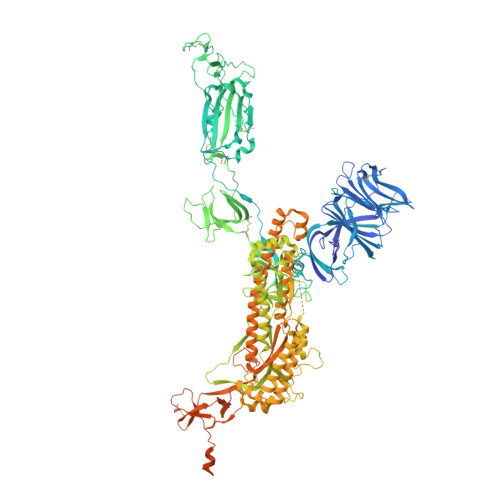
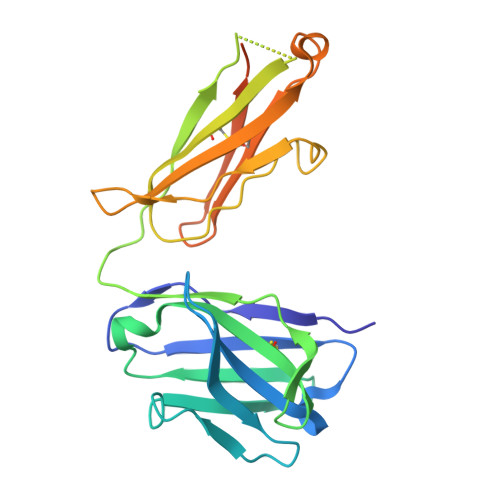
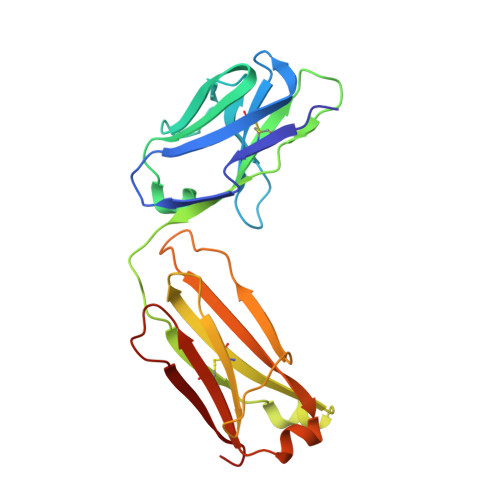
Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Zhu, X., Mannar, D., Srivastava, S.S., Berezuk, A.M., Demers, J.P., Saville, J.W., Leopold, K., Li, W., Dimitrov, D.S., Tuttle, K.S., Zhou, S., Chittori, S., Subramaniam, S.(2021) PLoS Biol 19: e3001237-e3001237
- PubMed: 33914735
- DOI: https://doi.org/10.1371/journal.pbio.3001237
- Primary Citation of Related Structures:
7MJG, 7MJH, 7MJI, 7MJJ, 7MJK, 7MJL, 7MJM, 7MJN - PubMed Abstract:
The recently reported "UK variant" (B.1.1.7) of SARS-CoV-2 is thought to be more infectious than previously circulating strains as a result of several changes, including the N501Y mutation. We present a 2.9-Å resolution cryo-electron microscopy (cryo-EM) structure of the complex between the ACE2 receptor and N501Y spike protein ectodomains that shows Y501 inserted into a cavity at the binding interface near Y41 of ACE2. This additional interaction provides a structural explanation for the increased ACE2 affinity of the N501Y mutant, and likely contributes to its increased infectivity. However, this mutation does not result in large structural changes, enabling important neutralization epitopes to be retained in the spike receptor binding domain. We confirmed this through biophysical assays and by determining cryo-EM structures of spike protein ectodomains bound to 2 representative potent neutralizing antibody fragments.
- Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia, Canada.
Organizational Affiliation: