Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Phillips, R.S., Ting, S.C., Anderson, K.(2021) Biochemistry 60: 1609-1618
- PubMed: 33949189
- DOI: https://doi.org/10.1021/acs.biochem.1c00106
- Primary Citation of Related Structures:
7LC0, 7LCE - PubMed Abstract:
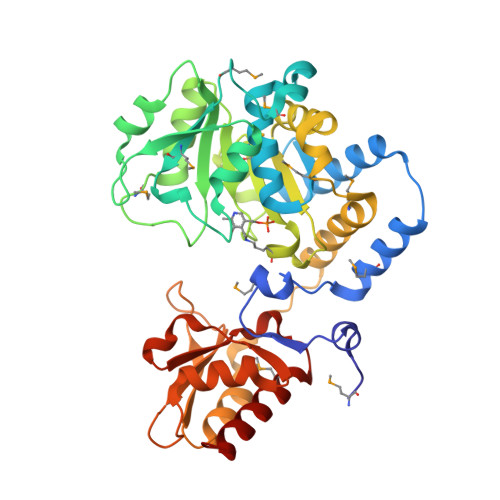
d-Glucosaminate-6-phosphate ammonia-lyase (DGL) is a pyridoxal 5'-phosphate (PLP)-dependent enzyme that produces 2-keto-3-deoxygluconate 6-phosphate (KDG-6-P) in the metabolism of d-glucosaminic acid by Salmonella enterica serovar typhimurium. We have determined the crystal structure of DGL by SAD phasing with selenomethionine to a resolution of 2.58 Å. The sequence has very low identity with most other members of the aminotransferase (AT) superfamily. The structure forms an octameric assembly as a tetramer of dimers that has not been observed previously in the AT superfamily. PLP is covalently bound as a Schiff base to Lys-213 in the catalytic dimer at the interface of two monomers. The structure lacks the conserved arginine that binds the α-carboxylate of the substrate in most members of the AT superfamily. However, there is a cluster of arginines in the small domain that likely serves as a binding site for the phosphate of the substrate. The deamination reaction performed in D 2 O gives a KDG-6-P product stereospecifically deuterated at C3; thus, the mechanism must involve an enamine intermediate that is protonated by the enzyme before product release. Nuclear magnetic resonance (NMR) analysis demonstrates that the deuterium is located in the pro - R position in the product, showing that the elimination of water takes place with inversion of configuration at C3, which is unprecedented for a PLP-dependent dehydratase/deaminase. On the basis of the crystal structure and the NMR data, a reaction mechanism for DGL is proposed.
- Department of Chemistry, University of Georgia, Athens, Georgia 30602, United States.
Organizational Affiliation: