Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Tarasova, E., Dhindwal, S., Popp, M., Hussain, S., Khayat, R.(2021) mBio 12: e0076321-e0076321
- PubMed: 34311576
- DOI: https://doi.org/10.1128/mBio.00763-21
- Primary Citation of Related Structures:
7LAR, 7LAS - PubMed Abstract:
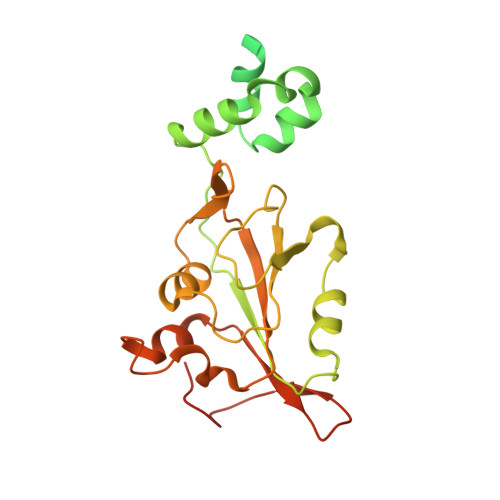
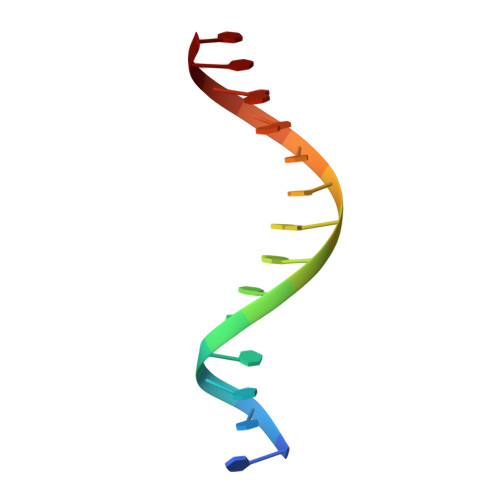
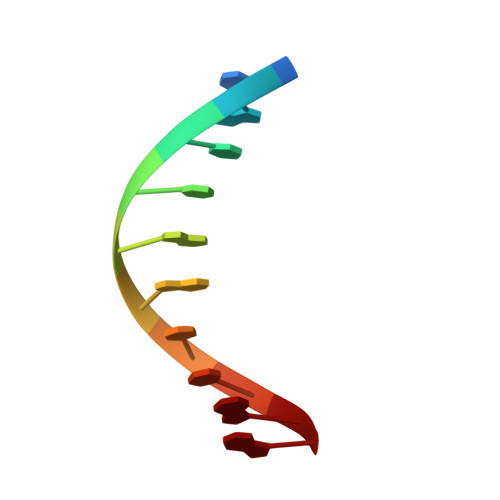
Circular Rep-encoding single-stranded DNA (CRESS-DNA) viruses infect members from all three domains of life ( Archaea , Prokarya , and Eukarya ). The replicase (Rep) from these viruses is responsible for initiating rolling circle replication (RCR) of their genomes. Rep is a multifunctional enzyme responsible for nicking and ligating ssDNA and unwinding double-stranded DNA (dsDNA). We report the structure of porcine circovirus 2 (PCV2) Rep bound to ADP and single-stranded DNA (ssDNA), and Rep bound to ADP and double-stranded DNA (dsDNA). The structures demonstrate Rep to be a member of the superfamily 3 (SF3) of ATPases Associated with diverse cellular Activities (AAA + ) superfamily clade 4. At the Rep N terminus is an endonuclease domain ( ED ) that is responsible for ssDNA nicking and ligation, in the center of Rep is an oligomerization domain ( OD ) responsible for hexamerization, and at the C terminus is an ATPase domain ( AD ) responsible for ssDNA/dsDNA interaction and translocation. The Rep AD binds to DNA such that the ED faces the replication fork. The six AD spiral around the DNA to interact with the backbone phosphates from four consecutive nucleotides. Three of the six AD are able to sense the backbone phosphates from the second strand of dsDNA. Heterogeneous classification of the data demonstrates the ED and AD to be mobile. Furthermore, we demonstrate that Rep exhibits basal nucleoside triphosphatase (NTPase) activity. IMPORTANCE CRESS-DNA viruses encompass a significant portion of the biosphere's virome. However, little is known about the structure of Rep responsible for initiating the RCR of CRESS-DNA viruses. We use cryo-electron microscopy (cryo-EM) to determine the structure of PCV2 Rep in complex with ADP and ss/dsDNA. Our structures demonstrate CRESS-DNA Reps to be SF3 members (clade 4) of the AAA+ superfamily. The structures further provide the mechanism by which CRESS-DNA virus Reps recognize DNA and translocate DNA for genome replication. Our structures also demonstrate the ED and AD of PCV2 Rep to be highly mobile. We propose the mobile nature of these domains to be necessary for proper functioning of Reps. We further demonstrate that Reps exhibit basal NTPase activity. Our studies also provide initial insight into the mechanism of RCR.
- Department of Chemistry and Biochemistry, The City College of New Yorkgrid.254250.4, New York, New York, USA.
Organizational Affiliation: