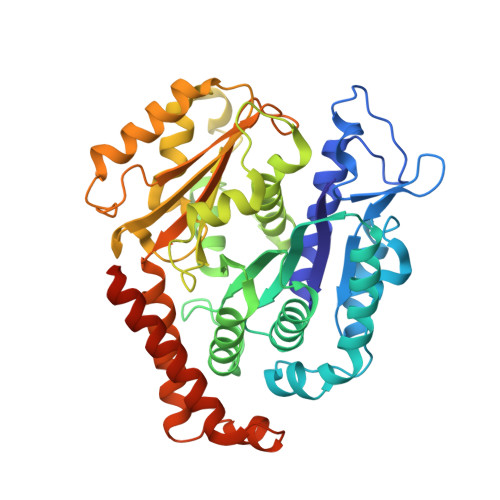
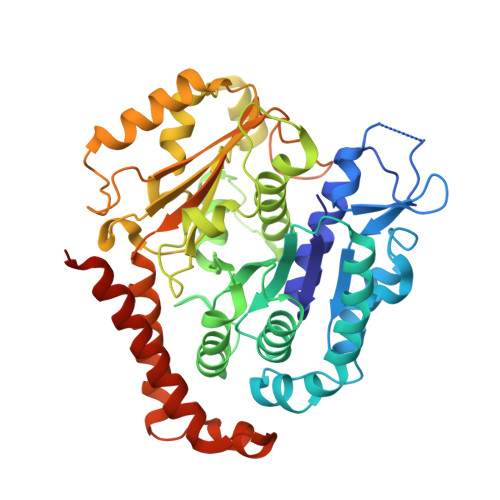
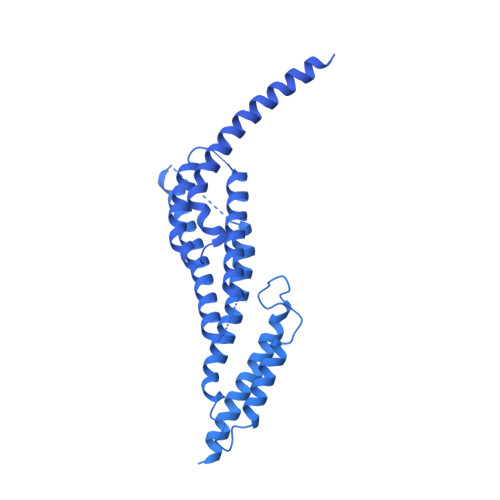
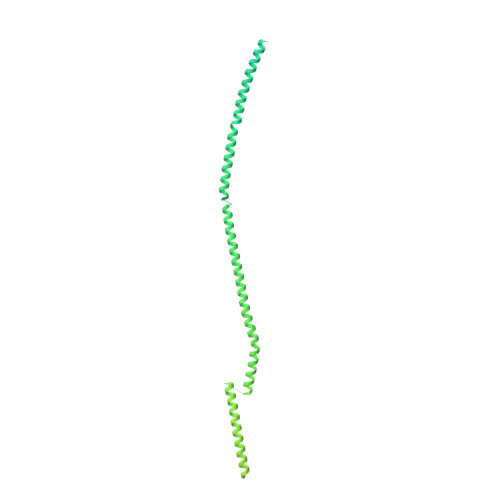
Structure of a microtubule-bound axonemal dynein.
Walton, T., Wu, H., Brown, A.(2021) Nat Commun 12: 477-477
- PubMed: 33473120
- DOI: https://doi.org/10.1038/s41467-020-20735-7
- Primary Citation of Related Structures:
7KZM, 7KZN, 7KZO - PubMed Abstract:
Axonemal dyneins are tethered to doublet microtubules inside cilia to drive ciliary beating, a process critical for cellular motility and extracellular fluid flow. Axonemal dyneins are evolutionarily and biochemically distinct from cytoplasmic dyneins that transport cargo, and the mechanisms regulating their localization and function are poorly understood. Here, we report a single-particle cryo-EM reconstruction of a three-headed axonemal dynein natively bound to doublet microtubules isolated from cilia. The slanted conformation of the axonemal dynein causes interaction of its motor domains with the neighboring dynein complex. Our structure shows how a heterotrimeric docking complex specifically localizes the linear array of axonemal dyneins to the doublet microtubule by directly interacting with the heavy chains. Our structural analysis establishes the arrangement of conserved heavy, intermediate and light chain subunits, and provides a framework to understand the roles of individual subunits and the interactions between dyneins during ciliary waveform generation.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, 240 Longwood Avenue, Boston, MA, USA.
Organizational Affiliation: