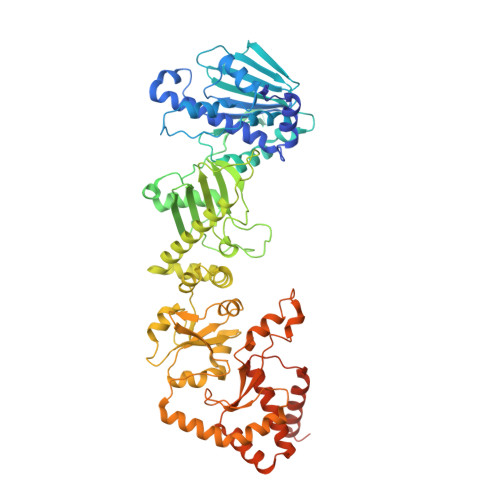
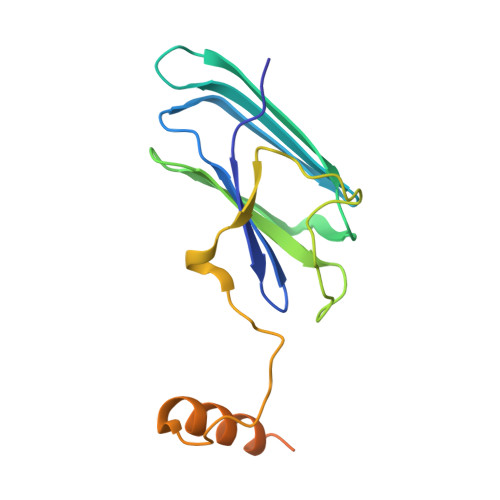
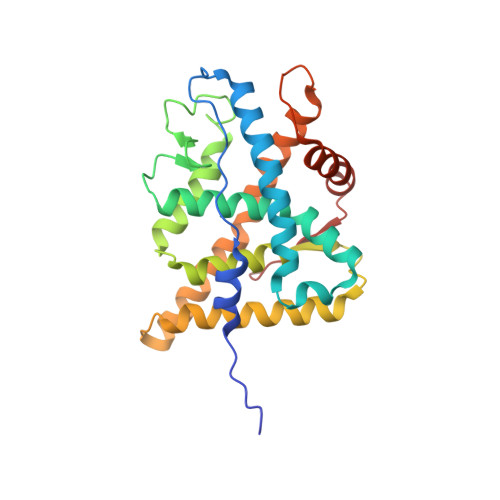
Structure of Hsp90-p23-GR reveals the Hsp90 client-remodelling mechanism.
Noddings, C.M., Wang, R.Y., Johnson, J.L., Agard, D.A.(2021) Nature 601: 465-469
- PubMed: 34937936
- DOI: https://doi.org/10.1038/s41586-021-04236-1
- Primary Citation of Related Structures:
7KRJ - PubMed Abstract:
Hsp90 is a conserved and essential molecular chaperone responsible for the folding and activation of hundreds of 'client' proteins 1-3 . The glucocorticoid receptor (GR) is a model client that constantly depends on Hsp90 for activity 4-9 . GR ligand binding was previously shown to nr inhibited by Hsp70 and restored by Hsp90, aided by the co-chaperone p23 10 . However, a molecular understanding of the chaperone-mediated remodelling that occurs between the inactive Hsp70-Hsp90 'client-loading complex' and an activated Hsp90-p23 'client-maturation complex' is lacking for any client, including GR. Here we present a cryo-electron microscopy (cryo-EM) structure of the human GR-maturation complex (GR-Hsp90-p23), revealing that the GR ligand-binding domain is restored to a folded, ligand-bound conformation, while being simultaneously threaded through the Hsp90 lumen. In addition, p23 directly stabilizes native GR using a C-terminal helix, resulting in enhanced ligand binding. This structure of a client bound to Hsp90 in a native conformation contrasts sharply with the unfolded kinase-Hsp90 structure 11 . Thus, aided by direct co-chaperone-client interactions, Hsp90 can directly dictate client-specific folding outcomes. Together with the GR-loading complex structure 12 , we present the molecular mechanism of chaperone-mediated GR remodelling, establishing the first, to our knowledge, complete chaperone cycle for any Hsp90 client.
- Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, CA, USA.
Organizational Affiliation: