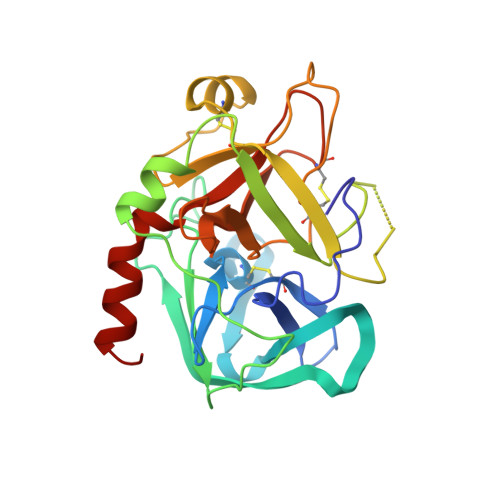
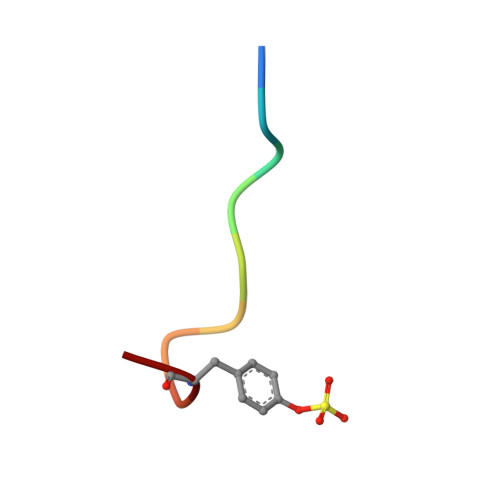
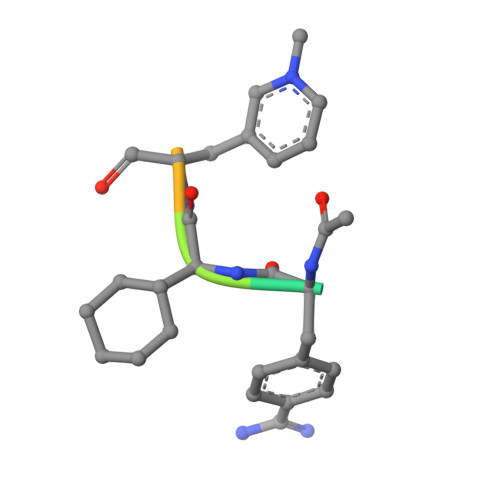
Structures of thrombin retro-inhibited with SEL2711 and SEL2770 as they relate to factor Xa binding.
Mochalkin, I., Tulinsky, A.(1999) Acta Crystallogr D Biol Crystallogr 55: 785-793
- PubMed: 10089309
- DOI: https://doi.org/10.1107/s0907444999000359
- Primary Citation of Related Structures:
7KME, 8KME - PubMed Abstract:
Most thrombin active-site inhibitors form a short antiparallel beta-strand with residues Ser214-Gly216. However, the Selectide Corp. inhibitors SEL2711 and SEL2770 bind to thrombin in a retro fashion, making a parallel beta-strand with Ser214-Gly216 similar to other retro-binding inhibitors. The crystallographic structures of thrombin-hirugen complexed with SEL2711 and SEL2770, which are isostructural with the binary thrombin-hirugen complex, have been determined and refined in the 9.0-2.1 A resolution range to final R values of 16.5 and 16.7%, respectively. The structures of the SEL2711 and SEL2770 complexes contain 131 and 104 water molecules, respectively, both of which correspond to occupancies of greater than 0.5. The L-4-amidinophenylalanyl residues of SEL2711 and SEL2770 are fixed at the S1 specificity site, utilizing favorable ionic and hydrogen-bonding interactions between the N atoms of the amidino group and the side-chain O atoms of Asp189. The Glu192 residue of thrombin adopts an extended conformation, which allows the L-cyclohexylglycyl residue in the P2 retro-binding position of the inhibitors to occupy a similar site to the P3 aspartate in thrombin platelet-receptor peptides bound to thrombin. The N-terminal acetyl group of both inhibitors is located in the S2 subsite, while the L-3-pyridyl-(3-methyl)-alanyl of SEL2711 and the L-(N,N-dimethyl)lysine of SEL2770 occupy the S3 D-Phe subsite of D-PheProArg chloromethyl ketone (PPACK) in the thrombin-PPACK complex. The two C-terminal residues of SEL2711 (leucine and proline) point into the solvent and have no electron density in the thrombin complex. Those of SEL2770 are also positioned into the solvent, but surprisingly produce weak electron density with high B values ( = 50 A2). Since the Selectide inhibitors are about 10(4) times more specific for factor Xa, modeling retro-binding to the latter suggests that the selectivity can be a consequence of interactions of the inhibitors in the S3-S4 binding subsites of factor Xa.
- Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, USA.
Organizational Affiliation: