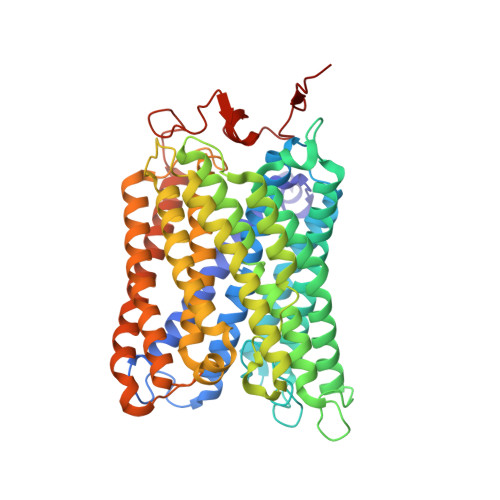
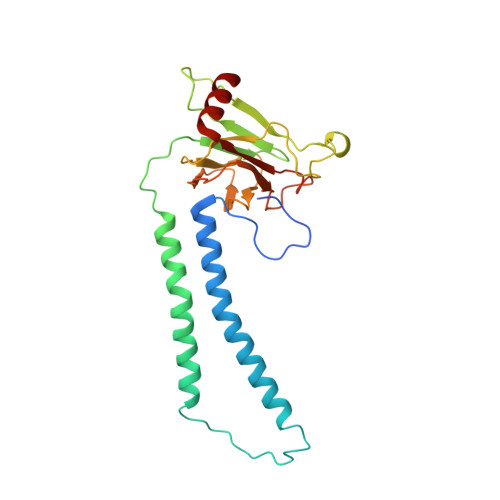
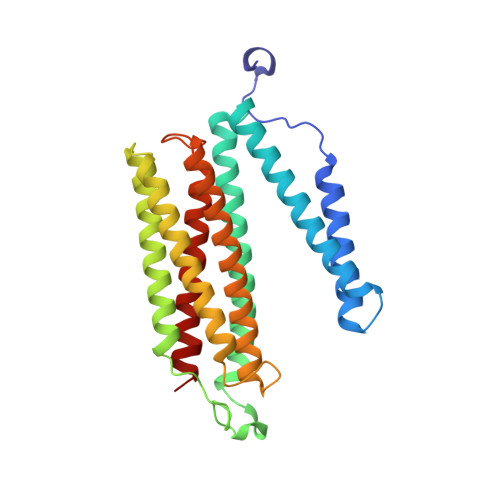
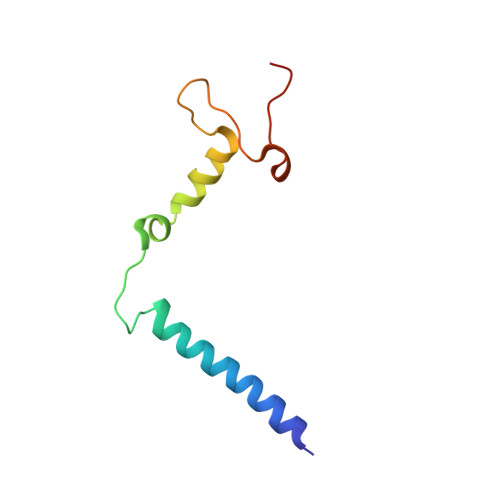
Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Maldonado, M., Guo, F., Letts, J.A.(2021) Elife 10
- PubMed: 33463523
- DOI: https://doi.org/10.7554/eLife.62047
- Primary Citation of Related Structures:
7JRG, 7JRO, 7JRP - PubMed Abstract:
Mitochondrial complex III (CIII 2 ) and complex IV (CIV), which can associate into a higher-order supercomplex (SC III 2 +IV), play key roles in respiration. However, structures of these plant complexes remain unknown. We present atomic models of CIII 2 , CIV, and SC III 2 +IV from Vigna radiata determined by single-particle cryoEM. The structures reveal plant-specific differences in the MPP domain of CIII 2 and define the subunit composition of CIV. Conformational heterogeneity analysis of CIII 2 revealed long-range, coordinated movements across the complex, as well as the motion of CIII 2 's iron-sulfur head domain. The CIV structure suggests that, in plants, proton translocation does not occur via the H channel. The supercomplex interface differs significantly from that in yeast and bacteria in its interacting subunits, angle of approach and limited interactions in the mitochondrial matrix. These structures challenge long-standing assumptions about the plant complexes and generate new mechanistic hypotheses.
- Department of Molecular and Cellular Biology, University of California Davis, Davis, United States.
Organizational Affiliation: