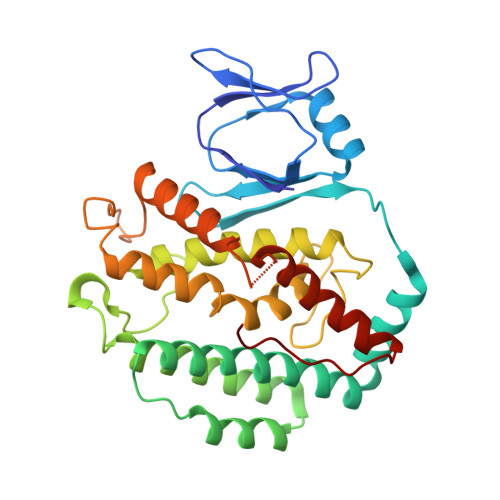
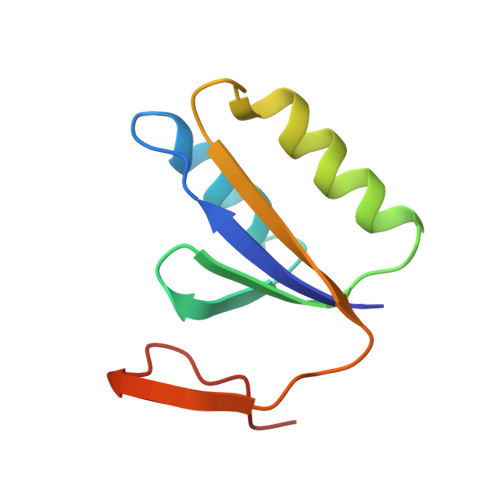
A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus.
Tang, D., Li, H., Wu, C., Jia, T., He, H., Yao, S., Yu, Y., Chen, Q.(2021) Int J Biol Macromol 183: 379-386
- PubMed: 33864868
- DOI: https://doi.org/10.1016/j.ijbiomac.2021.04.074
- Primary Citation of Related Structures:
7EI1 - PubMed Abstract:
In the adaptation stage of CRISPR-Cas systems, the Cas1-Cas2 integrase captures and integrates new invader-derived spacers into the CRISPR locus, serving as a molecular memory of prior infection. As of yet, the structural information of Cas1-Cas2 complex is available only for two species. Here we present the crystal structure of Cas1-Cas2 complex of Pyrococcus furiosus, which showed a distinct architecture from the known Cas1-Cas2 complexes. The shorter C-terminal tail of Pfu Cas2 directs the Cas1 dimers go in the opposite direction, resulting in a different prespacer binding mode. Based on our structural and mutagenesis results, we modeled a prespacer with a shorter duplex and longer 3' overhangs to bind Pfu Cas1-Cas2 complex. The prespacer preference was confirmed by EMSA, fluorescence polarization, and in vitro integration assays. This model provides a potential explanation for the longer spacer acquisition observed in P. furiosus when deleting both cas4 genes. Our study highlights the diversity of the CRISPR adaptation module.
- State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, PR China.
Organizational Affiliation: