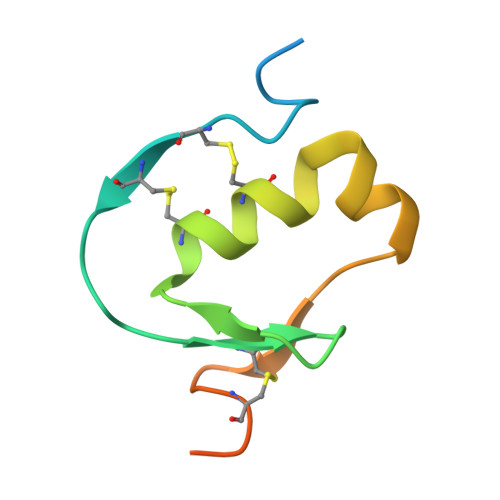
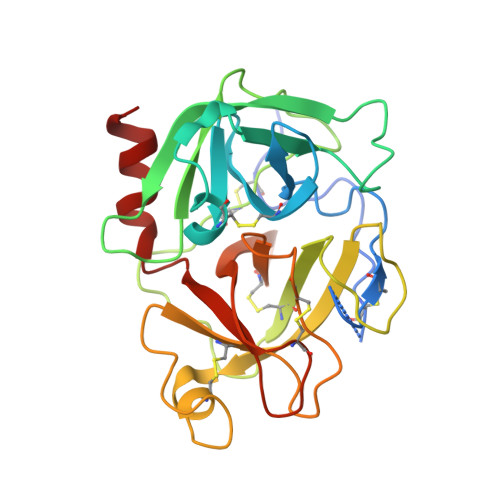
Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Walvekar, V.A., Ramesh, K., Jobichen, C., Kannan, M., Sivaraman, J., Kini, R.M., Mok, Y.K.(2022) Protein Sci 31: 470-484
- PubMed: 34800067
- DOI: https://doi.org/10.1002/pro.4245
- Primary Citation of Related Structures:
7E50 - PubMed Abstract:
Kazal-type protease inhibitor specificity is believed to be determined by sequence of the reactive-site loop that make most, if not all, contacts with the serine protease. Here, we determined the complex crystal structure of Aedes aegypti trypsin inhibitor (AaTI) with μ-plasmin, and compared its reactivities with other Kazal-type inhibitors, infestin-1 and infestin-4. We show that the shortened 99-loop of plasmin creates an S2 pocket, which is filled by phenylalanine at the P2 position of the reactive-site loop of infestin-4. In contrast, AaTI and infestin-1 retain a proline at P2, rendering the S2 pocket unfilled, which leads to lower plasmin inhibitions. Furthermore, the protein scaffold of AaTI is unstable, due to an elongated Cys-V to Cys-VI region leading to a less compact hydrophobic core. Chimeric study shows that the stability of the scaffold can be modified by swapping of this Cys-V to Cys-VI region between AaTI and infestin-4. The scaffold instability causes steric clashing of the bulky P2 residue, leading to significantly reduced inhibition of plasmin by AaTI or infestin-4 chimera. Our findings suggest that surface loops of protease and scaffold stability of Kazal-type inhibitor are both necessary for specific protease inhibition, in addition to reactive site loop sequence. PDB ID code: 7E50.
- Department of Biological Sciences, National University of Singapore, Singapore.
Organizational Affiliation: