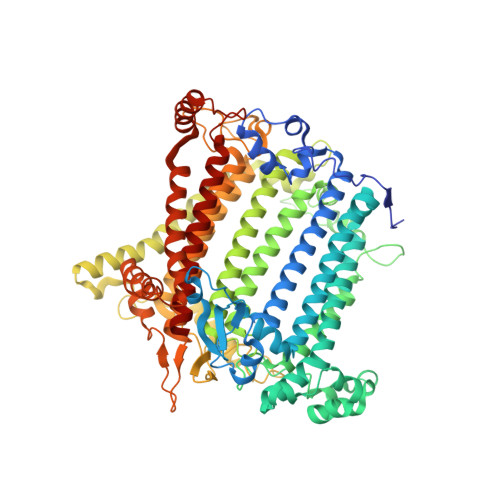
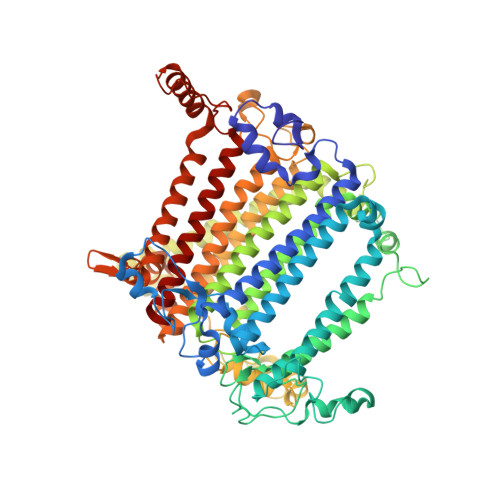
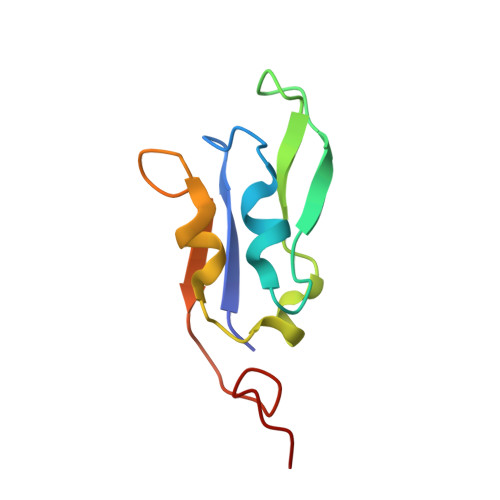
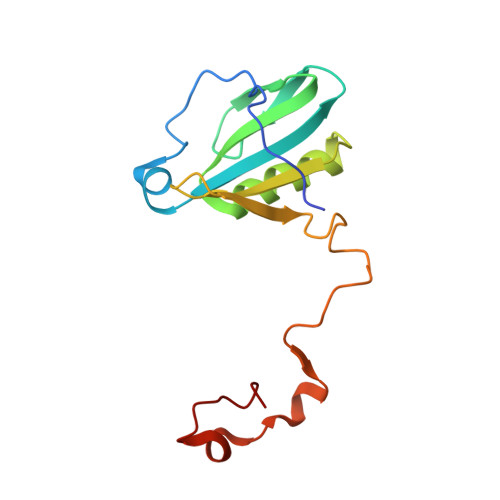
Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 angstrom resolution.
Wang, J., Yu, L.J., Wang, W., Yan, Q., Kuang, T., Qin, X., Shen, J.R.(2021) J Integr Plant Biol 63: 1367-1381
- PubMed: 33788400
- DOI: https://doi.org/10.1111/jipb.13095
- Primary Citation of Related Structures:
7DKZ - PubMed Abstract:
Photosystem I (PSI) is one of the two photosystems in photosynthesis, and performs a series of electron transfer reactions leading to the reduction of ferredoxin. In higher plants, PSI is surrounded by four light-harvesting complex I (LHCI) subunits, which harvest and transfer energy efficiently to the PSI core. The crystal structure of PSI-LHCI supercomplex has been analyzed up to 2.6 Å resolution, providing much information on the arrangement of proteins and cofactors in this complicated supercomplex. Here we have optimized crystallization conditions, and analyzed the crystal structure of PSI-LHCI at 2.4 Å resolution. Our structure showed some shift of the LHCI, especially the Lhca4 subunit, away from the PSI core, suggesting the indirect connection and inefficiency of energy transfer from this Lhca subunit to the PSI core. We identified five new lipids in the structure, most of them are located in the gap region between the Lhca subunits and the PSI core. These lipid molecules may play important roles in binding of the Lhca subunits to the core, as well as in the assembly of the supercomplex. The present results thus provide novel information for the elucidation of the mechanisms for the light-energy harvesting, transfer and assembly of this supercomplex.
- Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China.
Organizational Affiliation: