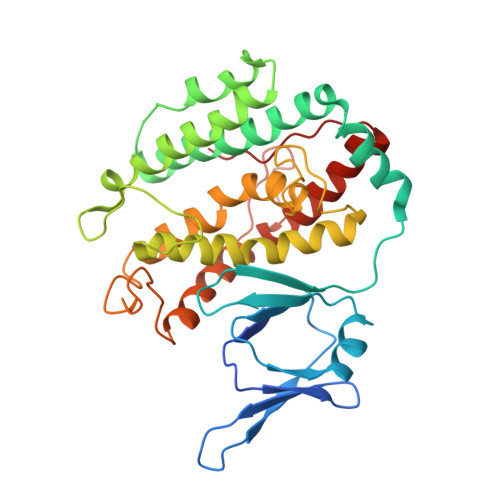
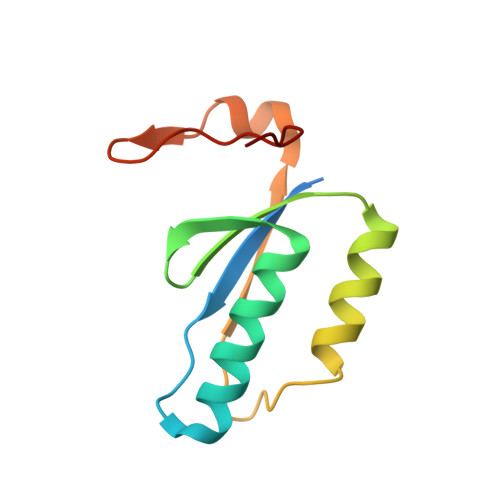
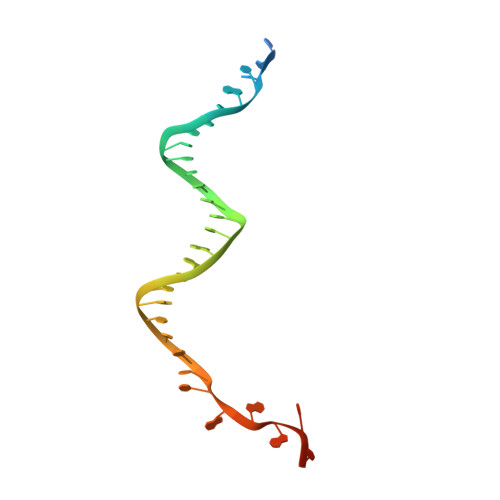
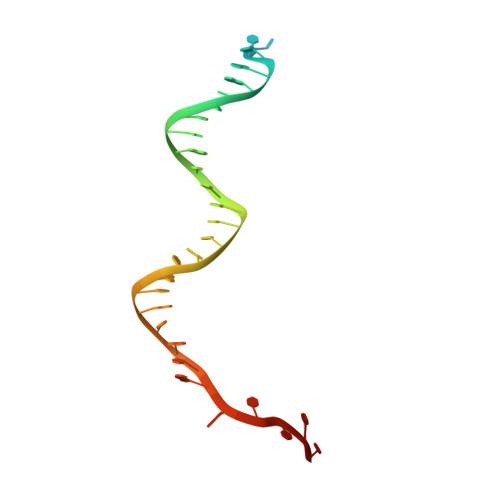
Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Wu, C., Tang, D., Cheng, J., Hu, D., Yang, Z., Ma, X., He, H., Yao, S., Fu, T.M., Yu, Y., Chen, Q.(2021) Nucleic Acids Res 49: 2973-2984
- PubMed: 33619565
- DOI: https://doi.org/10.1093/nar/gkab105
- Primary Citation of Related Structures:
7CR6, 7CR8 - PubMed Abstract:
CRISPR-Cas immune systems process and integrate short fragments of DNA from new invaders as spacers into the host CRISPR locus to establish molecular memory of prior infection, which is also known as adaptation in the field. Some CRISPR-Cas systems rely on Cas1 and Cas2 to complete the adaptation process, which has been characterized in a few systems. In contrast, many other CRISPR-Cas systems require an additional factor of Cas4 for efficient adaptation, the mechanism of which remains less understood. Here we present biochemical reconstitution of the Synechocystis sp. PCC6803 type I-D adaptation system, X-ray crystal structures of Cas1-Cas2-prespacer complexes, and negative stained electron microscopy structure of the Cas4-Cas1 complex. Cas4 and Cas2 compete with each other to interact with Cas1. In the absence of prespacer, Cas4 but not Cas2 assembles with Cas1 into a very stable complex for processing the prespacer. Strikingly, the Cas1-prespacer complex develops a higher binding affinity toward Cas2 to form the Cas1-Cas2-prespacer ternary complex for integration. Together, we show a two-step sequential assembly mechanism for the type I-D adaptation module of Synechocystis, in which Cas4-Cas1 and Cas1-Cas2 function as two exclusive complexes for prespacer processing, capture, and integration.
- State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu 610041, P.R. China.
Organizational Affiliation: