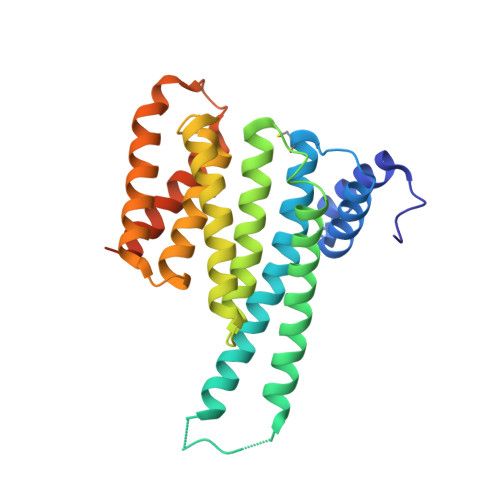
Fusicoccin-A Targets Cancerous Inhibitor of Protein Phosphatase 2A by Stabilizing a C-Terminal Interaction with 14-3-3.
Brink, H.J., van Senten, J.R., De Vries-van Leeuwen, I.J., da Costa Pereira, D., Piersma, S.R., Jimenez, C.R., Centorrino, F., Ottmann, C., Siderius, M., Smit, M.J., de Boer, A.H.(2022) ACS Chem Biol 17: 2972-2978
- PubMed: 36255265
- DOI: https://doi.org/10.1021/acschembio.2c00299
- Primary Citation of Related Structures:
7BM9, 7BMC - PubMed Abstract:
The cancerous inhibitor of protein phosphatase 2A (CIP2A) is an oncoprotein found overexpressed in many types of cancer. CIP2A has been shown to stabilize oncoproteins such as cMYC by shielding them from PP2A-mediated dephosphorylation. Here we report that the penultimate residue Ser904 in the C-terminus of CIP2A can be phosphorylated to create a binding site for the regulatory protein 14-3-3. We demonstrate that 14-3-3 is a new interaction partner of CIP2A. The 14-3-3/CIP2A C-terminal interaction complex can be targeted by the protein-protein interaction (PPI) stabilizer fusicoccin-A (FC-A), resulting in enhanced levels of phosphorylated Ser904. FC-A treatment of TNBC cells leads to the increased association of CIP2A with 14-3-3. We show that the composite interface between 14 and 3-3 and CIP2A's C-terminus can be targeted by the PPI stabilizer FC-A, providing a new interface that could potentially be exploited to modulate CIP2A's activity.
- Amsterdam Institute for Molecular and Life Sciences (AIMMS), Division of Medicinal Chemistry, Faculty of Sciences, Vrije Universiteit, De Boelelaan 1108, Amsterdam 1081 HZ, The Netherlands.
Organizational Affiliation: