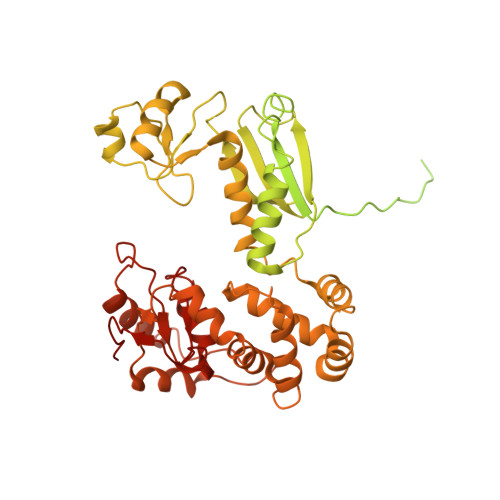
Nonameric structures of the cytoplasmic domain of FlhA and SctV in the context of the full-length protein.
Kuhlen, L., Johnson, S., Cao, J., Deme, J.C., Lea, S.M.(2021) PLoS One 16: e0252800-e0252800
- PubMed: 34143799
- DOI: https://doi.org/10.1371/journal.pone.0252800
- Primary Citation of Related Structures:
7ALW, 7AMY - PubMed Abstract:
Type three secretion is the mechanism of protein secretion found in bacterial flagella and injectisomes. At its centre is the export apparatus (EA), a complex of five membrane proteins through which secretion substrates pass the inner membrane. While the complex formed by four of the EA proteins has been well characterised structurally, little is known about the structure of the membrane domain of the largest subunit, FlhA in flagella, SctV in injectisomes. Furthermore, the biologically relevant nonameric assembly of FlhA/SctV has been infrequently observed and differences in conformation of the cytoplasmic portion of FlhA/SctV between open and closed states have been suggested to reflect secretion system specific differences. FlhA has been shown to bind to chaperone-substrate complexes in an open state, but in previous assembled ring structures, SctV is in a closed state. Here, we identify FlhA and SctV homologues that can be recombinantly produced in the oligomeric state and study them using cryo-electron microscopy. The structures of the cytoplasmic domains from both FlhA and SctV are in the open state and we observe a conserved interaction between a short stretch of residues at the N-terminus of the cytoplasmic domain, known as FlhAL/SctVL, with a groove on the adjacent protomer's cytoplasmic domain, which stabilises the nonameric ring assembly.
- Sir William Dunn School of Pathology, Oxford, United Kingdom.
Organizational Affiliation: