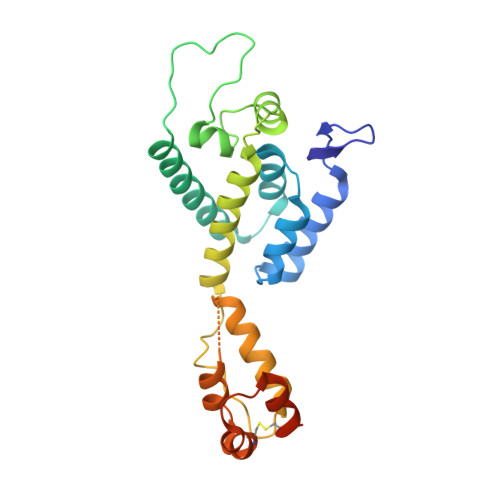
CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
Nicastro, G., Lucci, M., Oregioni, A., Kelly, G., Frenkiel, T.A., Taylor, I.A.(2022) J Mol Biology 434: 167691-167691
- PubMed: 35738429
- DOI: https://doi.org/10.1016/j.jmb.2022.167691
- Primary Citation of Related Structures:
7ZUD - PubMed Abstract:
Solution and solid-state NMR spectroscopy are highly complementary techniques for studying structure and dynamics in very high molecular weight systems. Here we have analysed the dynamics of HIV-1 capsid (CA) assemblies in presence of the cofactors IP6 and ATPγS and the host-factor CPSF6 using a combination of solution state and cross polarisation magic angle spinning (CP-MAS) solid-state NMR. In particular, dynamical effects on ns to µs and µs to ms timescales are observed revealing diverse motions in assembled CA. Using CP-MAS NMR, we exploited the sensitivity of the amide/Cα-Cβ backbone chemical shifts in DARR and NCA spectra to observe the plasticity of the HIV-1 CA tubular assemblies and also map the binding of cofactors and the dynamics of cofactor-CA complexes. In solution, we measured how the addition of host- and co-factors to CA -hexamers perturbed the chemical shifts and relaxation properties of CA-Ile and -Met methyl groups using transverse-relaxation-optimized NMR spectroscopy to exploit the sensitivity of methyl groups as probes in high-molecular weight proteins. These data show how dynamics of the CA protein assembly over a range of spatial and temporal scales play a critical role in CA function. Moreover, we show that binding of IP6, ATPγS and CPSF6 results in local chemical shift as well as dynamic changes for a significant, contiguous portion of CA, highlighting how allosteric pathways communicate ligand interactions between adjacent CA protomers.
- Macromolecular Structure Laboratory, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.
Organizational Affiliation: