Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
Vance, N.R., Ayala, R., Willhoft, O., Tvardovskiy, A., McCormack, E.A., Bartke, T., Zhang, X., Wigley, D.B.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RuvB-like 1 | 456 | Homo sapiens | Mutation(s): 0 Gene Names: RUVBL1, INO80H, NMP238, TIP49, TIP49A EC: 3.6.4.12 | 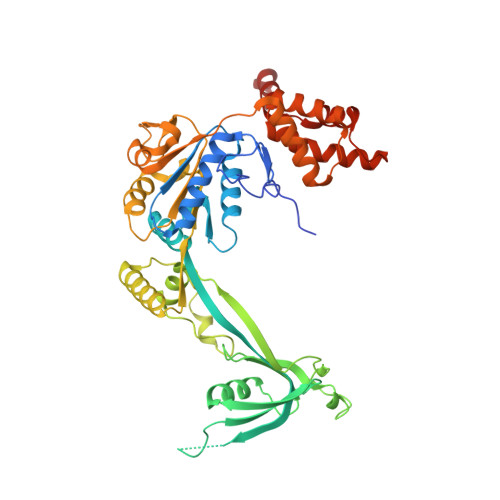 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y265 (Homo sapiens) Explore Q9Y265 Go to UniProtKB: Q9Y265 | |||||
PHAROS: Q9Y265 GTEx: ENSG00000175792 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y265 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RuvB-like 2 | 463 | Homo sapiens | Mutation(s): 0 Gene Names: RUVBL2, INO80J, TIP48, TIP49B, CGI-46 EC: 3.6.4.12 | 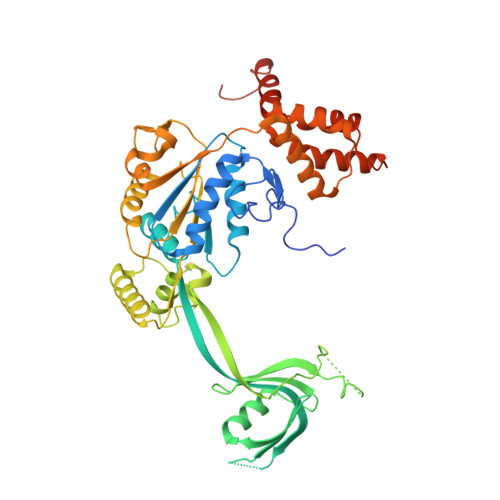 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y230 (Homo sapiens) Explore Q9Y230 Go to UniProtKB: Q9Y230 | |||||
PHAROS: Q9Y230 GTEx: ENSG00000183207 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y230 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chromatin-remodeling ATPase INO80 | 1,556 | Homo sapiens | Mutation(s): 0 Gene Names: INO80, INO80A, INOC1, KIAA1259 EC: 3.6.4 | 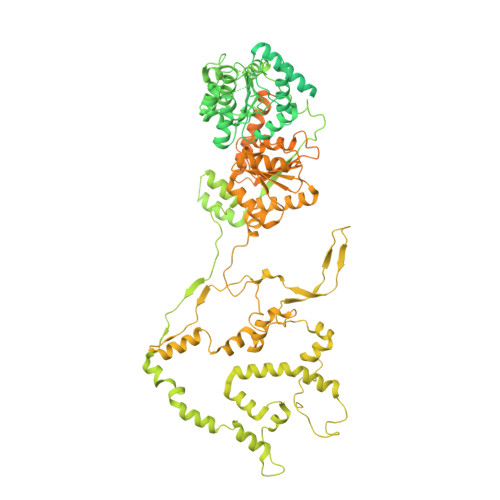 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9ULG1 (Homo sapiens) Explore Q9ULG1 Go to UniProtKB: Q9ULG1 | |||||
PHAROS: Q9ULG1 GTEx: ENSG00000128908 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ULG1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | H [auth N], M [auth J] | 103 | Homo sapiens | Mutation(s): 0 Gene Names: | 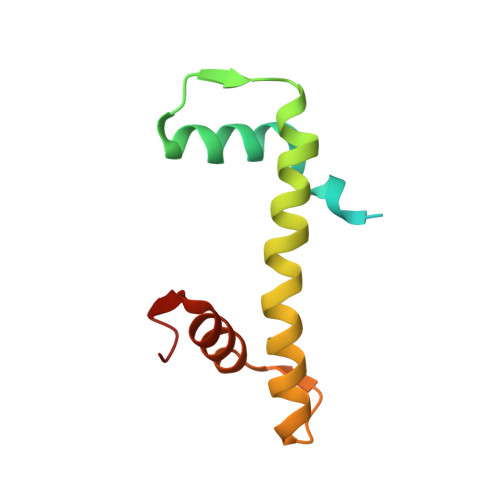 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62805 (Homo sapiens) Explore P62805 Go to UniProtKB: P62805 | |||||
PHAROS: P62805 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62805 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| INO80 complex subunit B | I [auth R] | 356 | Homo sapiens | Mutation(s): 0 Gene Names: INO80B, HMGA1L4, PAPA1, ZNHIT4 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9C086 (Homo sapiens) Explore Q9C086 Go to UniProtKB: Q9C086 | |||||
GTEx: ENSG00000115274 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9C086 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 5 | J [auth H] | 607 | Homo sapiens | Mutation(s): 0 Gene Names: ACTR5, ARP5 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9H9F9 (Homo sapiens) Explore Q9H9F9 Go to UniProtKB: Q9H9F9 | |||||
PHAROS: Q9H9F9 GTEx: ENSG00000101442 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9H9F9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| INO80 complex subunit C | K [auth Q] | 192 | Homo sapiens | Mutation(s): 0 Gene Names: INO80C, C18orf37 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6PI98 (Homo sapiens) Explore Q6PI98 Go to UniProtKB: Q6PI98 | |||||
PHAROS: Q6PI98 GTEx: ENSG00000153391 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PI98 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.1 | L [auth I], P [auth M] | 136 | Homo sapiens | Mutation(s): 0 Gene Names: |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P68431 (Homo sapiens) Explore P68431 Go to UniProtKB: P68431 | |||||
PHAROS: P68431 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68431 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A type 1-B/E | N [auth K], Q [auth O] | 130 | Homo sapiens | Mutation(s): 0 Gene Names: HIST1H2AB, H2AFM, HIST1H2AE, H2AFA | 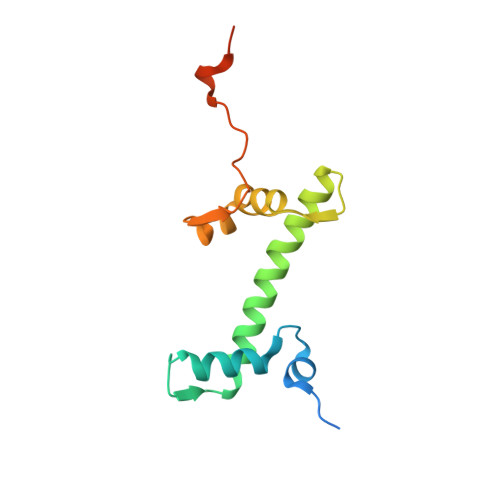 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04908 (Homo sapiens) Explore P04908 Go to UniProtKB: P04908 | |||||
PHAROS: P04908 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04908 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 1-J | O [auth L], R [auth P] | 126 | Homo sapiens | Mutation(s): 0 Gene Names: HIST1H2BJ, H2BFR | 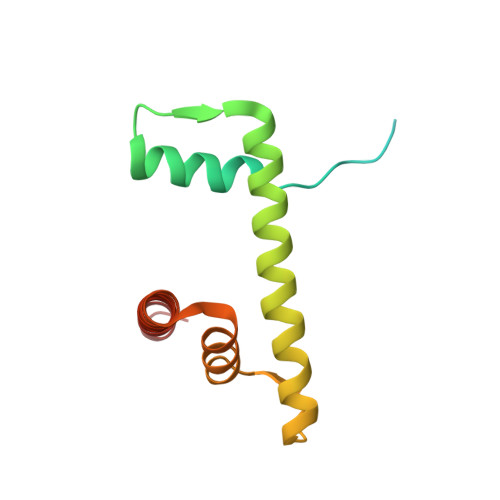 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06899 (Homo sapiens) Explore P06899 Go to UniProtKB: P06899 | |||||
PHAROS: P06899 GTEx: ENSG00000124635 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (158-MER) | S [auth X] | 158 | synthetic construct | 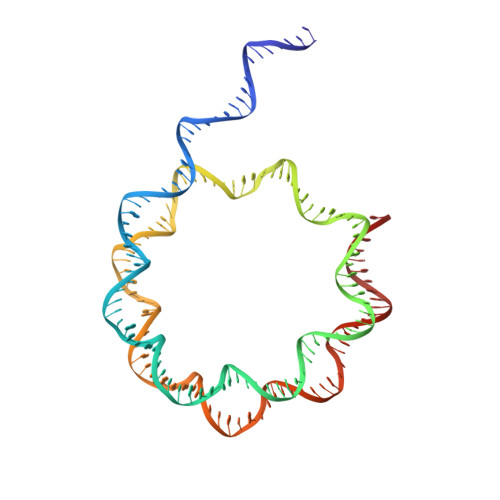 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (158-MER) | T [auth Y] | 158 | synthetic construct | 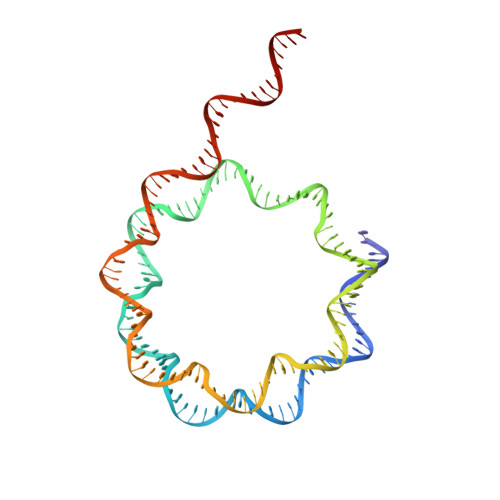 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | AA [auth G] DA [auth H] U [auth A] V [auth B] W [auth C] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| BEF (Subject of Investigation/LOI) Query on BEF | BA [auth G] | BERYLLIUM TRIFLUORIDE ION Be F3 OGIAHMCCNXDTIE-UHFFFAOYSA-K |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | CA [auth R] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 4 |
| MODEL REFINEMENT | PHENIX | 1.19.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 209327/Z/17/Z |