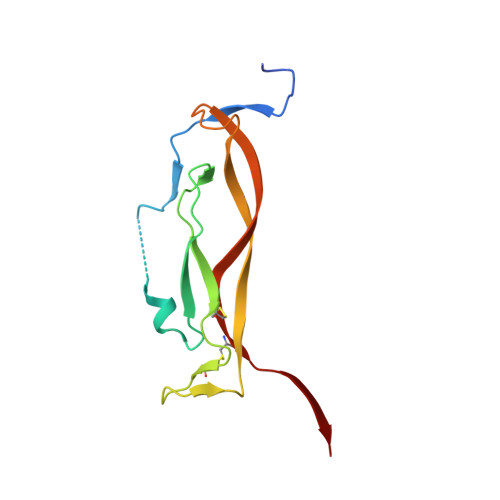
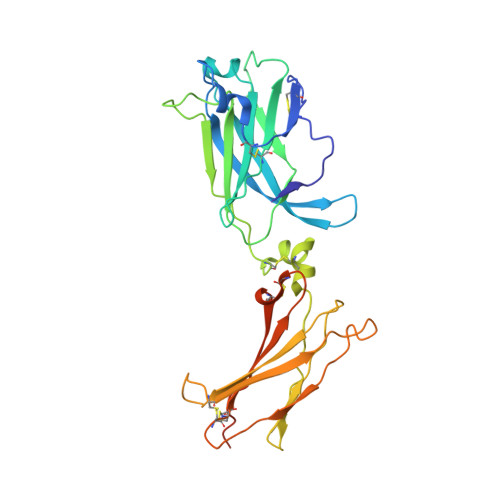
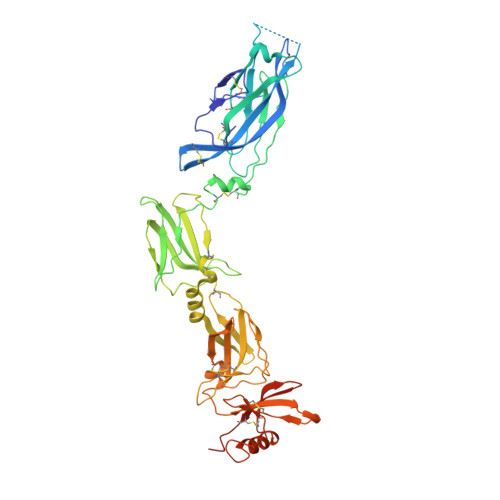
IL-17-induced dimerization of IL-17RA drives the formation of the IL-17 signalosome to potentiate signaling.
Goepfert, A., Barske, C., Lehmann, S., Wirth, E., Willemsen, J., Gudjonsson, J.E., Ward, N.L., Sarkar, M.K., Hemmig, R., Kolbinger, F., Rondeau, J.M.(2022) Cell Rep 41: 111489-111489
- PubMed: 36260993
- DOI: https://doi.org/10.1016/j.celrep.2022.111489
- Primary Citation of Related Structures:
5N9B, 7ZAN - PubMed Abstract:
Signaling through innate immune receptors such as the Toll-like receptor (TLR)/interleukin-1 receptor (IL-1R) superfamily proceeds via the assembly of large membrane-proximal complexes or "signalosomes." Although structurally distinct, the IL-17 receptor family triggers cellular responses that are typical of innate immune receptors. The IL-17RA receptor subunit is shared by several members of the IL-17 family. Using a combination of crystallographic, biophysical, and mutational studies, we show that IL-17A, IL-17F, and IL-17A/F induce IL-17RA dimerization. X-ray analysis of the heteromeric IL-17A complex with the extracellular domains of the IL-17RA and IL-17RC receptors reveals that cytokine-induced IL-17RA dimerization leads to the formation of a 2:2:2 hexameric signaling assembly. Furthermore, we demonstrate that the formation of the IL-17 signalosome potentiates IL-17-induced IL-36γ and CXCL1 mRNA expression in human keratinocytes, compared with a dimerization-defective IL-17RA variant.
- Novartis Institutes for BioMedical Research, Novartis Pharma AG, 4002 Basel, Switzerland.
Organizational Affiliation: