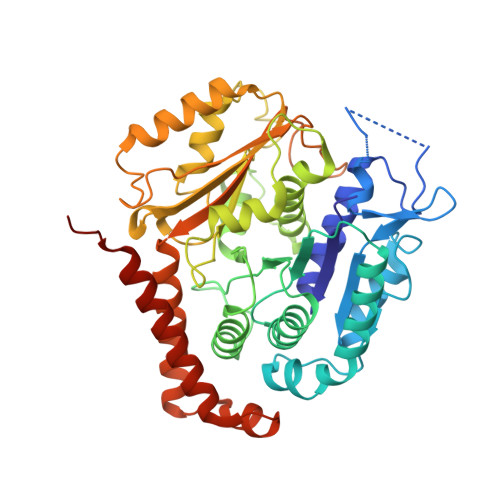
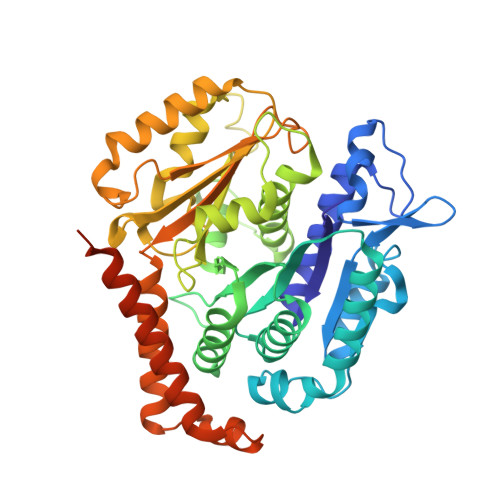
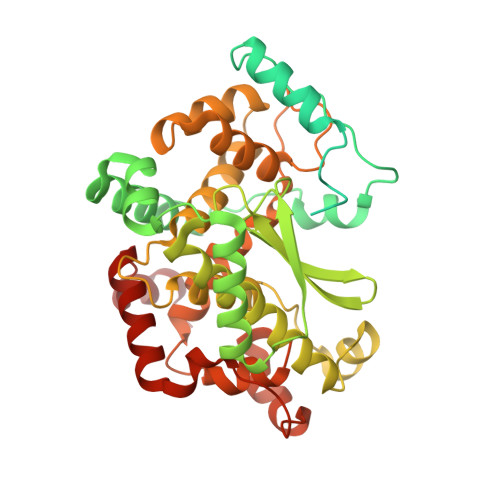
Posttranslational modification of microtubules by the MATCAP detyrosinase.
Landskron, L., Bak, J., Adamopoulos, A., Kaplani, K., Moraiti, M., van den Hengel, L.G., Song, J.Y., Bleijerveld, O.B., Nieuwenhuis, J., Heidebrecht, T., Henneman, L., Moutin, M.J., Barisic, M., Taraviras, S., Perrakis, A., Brummelkamp, T.R.(2022) Science 376: eabn6020-eabn6020
- PubMed: 35482892
- DOI: https://doi.org/10.1126/science.abn6020
- Primary Citation of Related Structures:
7Z5G, 7Z5H, 7Z6S - PubMed Abstract:
The detyrosination-tyrosination cycle involves the removal and religation of the C-terminal tyrosine of α-tubulin and is implicated in cognitive, cardiac, and mitotic defects. The vasohibin-small vasohibin-binding protein (SVBP) complex underlies much, but not all, detyrosination. We used haploid genetic screens to identify an unannotated protein, microtubule associated tyrosine carboxypeptidase (MATCAP), as a remaining detyrosinating enzyme. X-ray crystallography and cryo-electron microscopy structures established MATCAP's cleaving mechanism, substrate specificity, and microtubule recognition. Paradoxically, whereas abrogation of tyrosine religation is lethal in mice, codeletion of MATCAP and SVBP is not. Although viable, defective detyrosination caused microcephaly, associated with proliferative defects during neurogenesis, and abnormal behavior. Thus, MATCAP is a missing component of the detyrosination-tyrosination cycle, revealing the importance of this modification in brain formation.
- Oncode Institute, Division of Biochemistry, Netherlands Cancer Institute, 1066CX Amsterdam, Netherlands.
Organizational Affiliation: