Crystal structure DYRK1A in complex with CX-4945
Grygier, P., Pustelny, K., Golik, P., Popowicz, G., Dubin, G., Czarna, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1A | 370 | Homo sapiens | Mutation(s): 0 Gene Names: DYRK1A, DYRK, MNB, MNBH EC: 2.7.12.1 (PDB Primary Data), 2.7.11.23 (UniProt) | 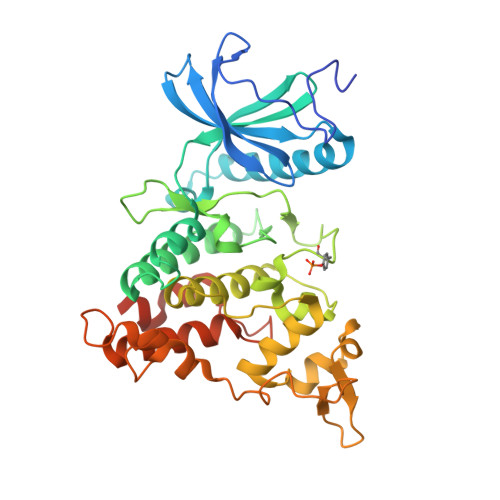 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13627 (Homo sapiens) Explore Q13627 Go to UniProtKB: Q13627 | |||||
PHAROS: Q13627 GTEx: ENSG00000157540 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13627 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 3NG Query on 3NG | CB [auth G] GA [auth C] I [auth A] JB [auth H] OA [auth D] | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid C19 H12 Cl N3 O2 MUOKSQABCJCOPU-UHFFFAOYSA-N |  | ||
| PG4 Query on PG4 | AA [auth B] BA [auth B] KA [auth C] LB [auth H] O [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | FA [auth B], MB [auth H] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | FB [auth G], GB [auth G], Q [auth A], XA [auth E] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | AB [auth F] DB [auth G] EB [auth G] HA [auth C] IA [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | BB [auth F] CA [auth B] DA [auth B] EA [auth B] HB [auth G] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PTR Query on PTR | A, B, C, D, E A, B, C, D, E, F, G, H | L-PEPTIDE LINKING | C9 H12 N O6 P |  | TYR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 247.682 | α = 90 |
| b = 134.317 | β = 96.48 |
| c = 121.717 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Polish National Science Centre | Poland | 2019/34/E/NZ1/00467 |
| National Center for Research and Development (Poland) | Poland | PPN/PPO/2018/1/00046 |