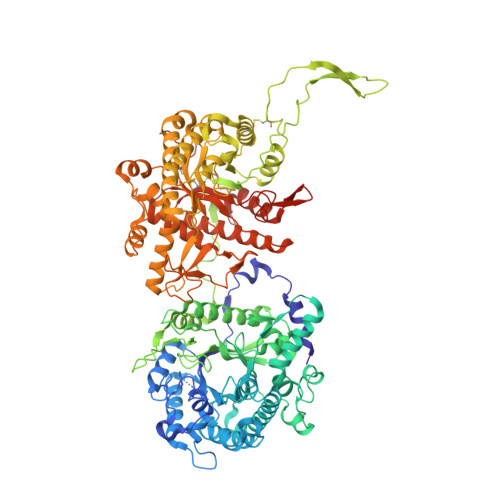
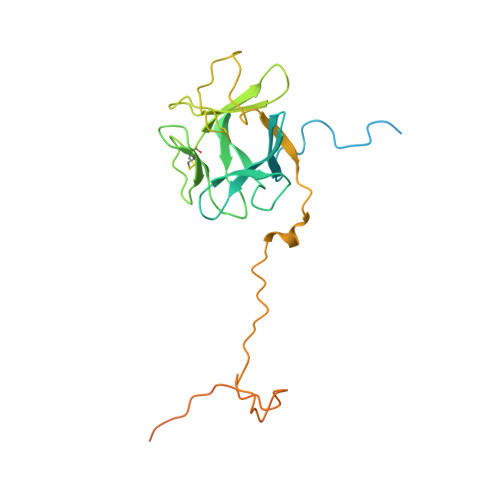
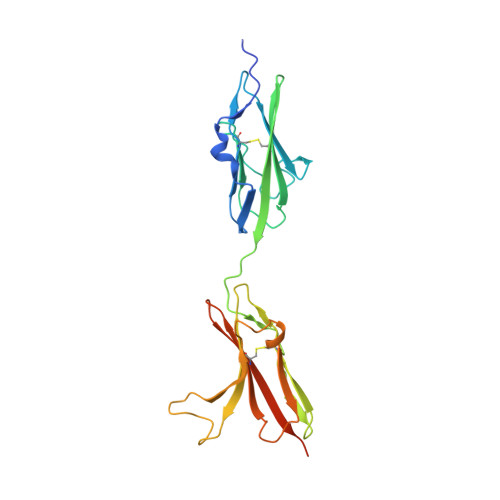
Structural basis for FGF hormone signalling.
Chen, L., Fu, L., Sun, J., Huang, Z., Fang, M., Zinkle, A., Liu, X., Lu, J., Pan, Z., Wang, Y., Liang, G., Li, X., Chen, G., Mohammadi, M.(2023) Nature 618: 862-870
- PubMed: 37286607
- DOI: https://doi.org/10.1038/s41586-023-06155-9
- Primary Citation of Related Structures:
7YSH, 7YSU, 7YSW - PubMed Abstract:
α/βKlotho coreceptors simultaneously engage fibroblast growth factor (FGF) hormones (FGF19, FGF21 and FGF23) 1,2 and their cognate cell-surface FGF receptors (FGFR1-4) thereby stabilizing the endocrine FGF-FGFR complex 3-6 . However, these hormones still require heparan sulfate (HS) proteoglycan as an additional coreceptor to induce FGFR dimerization/activation and hence elicit their essential metabolic activities 6 . To reveal the molecular mechanism underpinning the coreceptor role of HS, we solved cryo-electron microscopy structures of three distinct 1:2:1:1 FGF23-FGFR-αKlotho-HS quaternary complexes featuring the 'c' splice isoforms of FGFR1 (FGFR1c), FGFR3 (FGFR3c) or FGFR4 as the receptor component. These structures, supported by cell-based receptor complementation and heterodimerization experiments, reveal that a single HS chain enables FGF23 and its primary FGFR within a 1:1:1 FGF23-FGFR-αKlotho ternary complex to jointly recruit a lone secondary FGFR molecule leading to asymmetric receptor dimerization and activation. However, αKlotho does not directly participate in recruiting the secondary receptor/dimerization. We also show that the asymmetric mode of receptor dimerization is applicable to paracrine FGFs that signal solely in an HS-dependent fashion. Our structural and biochemical data overturn the current symmetric FGFR dimerization paradigm and provide blueprints for rational discovery of modulators of FGF signalling 2 as therapeutics for human metabolic diseases and cancer.
- Oujiang Laboratory (Zhejiang Lab for Regenerative Medicine, Vision, and Brain Health), School of Pharmaceutical Sciences, Wenzhou Medical University, Wenzhou, China.
Organizational Affiliation: