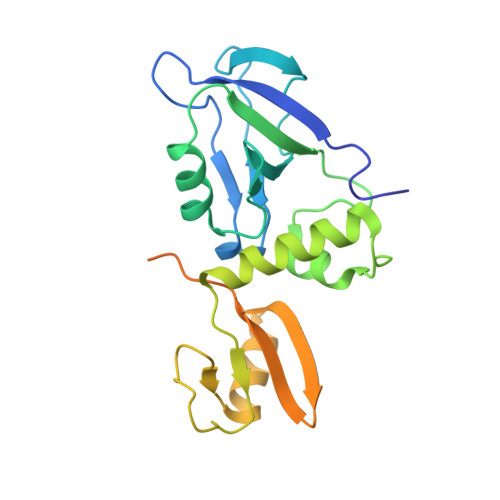
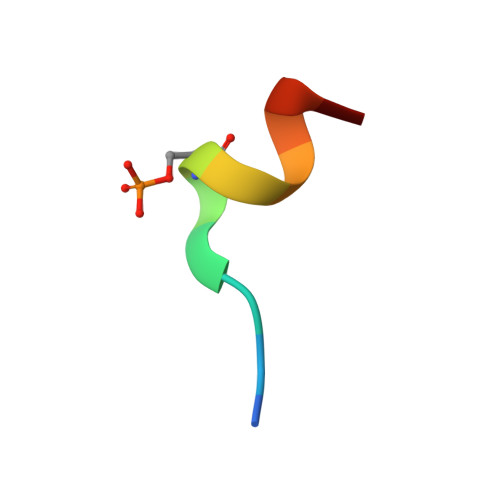Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Zhang, M., Cao, A., Lin, L., Chen, Y., Shang, Y., Wang, C., Zhang, M., Zhu, J.(2023) Sci Adv 9: eadf3295-eadf3295
- PubMed: 37163606
- DOI: https://doi.org/10.1126/sciadv.adf3295
- Primary Citation of Related Structures:
7YKF, 7YKG, 7YKH, 7YKI - PubMed Abstract:
Dynamic signal transduction requires the rapid assembly and disassembly of signaling complexes, often mediated by phosphoprotein binding modules. The guanylate kinase-like (GK) domain of the membrane-associated guanylate kinases (MAGUKs) is such a module orchestrating signaling at cellular junctions. The MAGI subfamily of MAGUKs contains a truncated GK domain with unknown structure and function, although they participate in diverse physiological and pathological processes. Here, we demonstrate that the truncated GK domain of MAGI2 interacts with its adjacent PDZ0 domain to form a structural supramodule capable of recognizing phosphoproteins. A conserved phosphorylation-dependent binding motif for PDZ0-GK is delineated, which leads to identification of a set of previously unknown binding partners. We explore the structure and function of the MAGI2-target complex with an inhibitory peptide derived from the consensus motif. Our work reveals an action mechanism of the cryptic MAGI GKs and broadens our understanding of the target recognition rules of phosphoprotein binding modules.
- Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders, Ministry of Education, Shanghai Jiao Tong University, Shanghai 200240, China.
Organizational Affiliation: