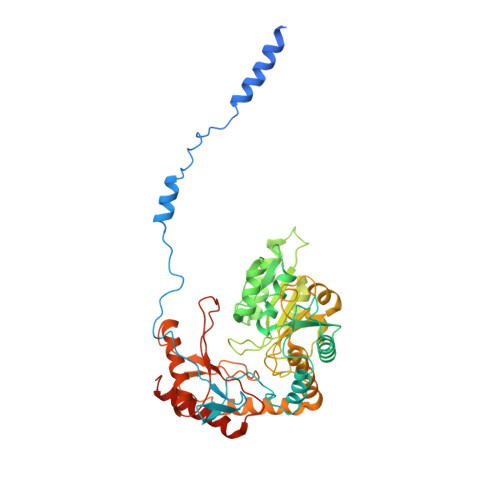
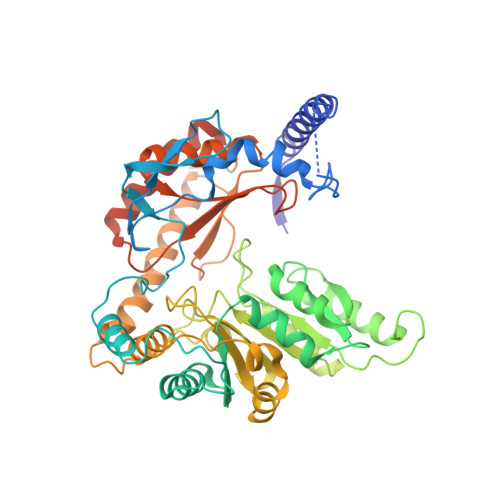
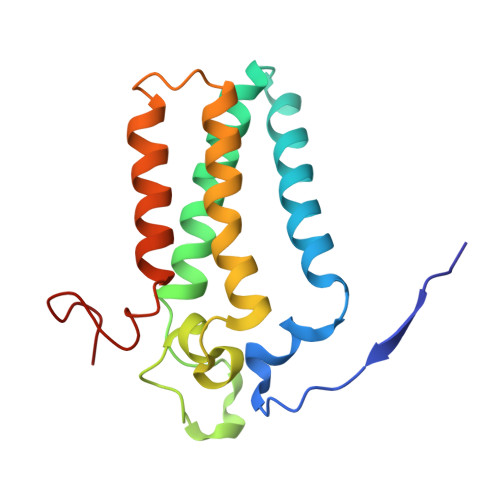
Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Liu, P., Xie, T., Wu, X., Han, G., Gupta, S.D., Zhang, Z., Yue, J., Dong, F., Gable, K., Niranjanakumari, S., Li, W., Wang, L., Liu, W., Yao, R., Cahoon, E.B., Dunn, T.M., Gong, X.(2023) Sci Adv 9: eadg0728-eadg0728
- PubMed: 36989369
- DOI: https://doi.org/10.1126/sciadv.adg0728
- Primary Citation of Related Structures:
7YJK, 7YJM, 7YJN, 7YJO - PubMed Abstract:
The serine palmitoyltransferase (SPT) complex catalyzes the first and rate-limiting step in sphingolipid biosynthesis in all eukaryotes. ORM/ORMDL proteins are negative regulators of SPT that respond to cellular sphingolipid levels. However, the molecular basis underlying ORM/ORMDL-dependent homeostatic regulation of SPT is not well understood. We determined the cryo-electron microscopy structure of Arabidopsis SPT-ORM1 complex, composed of LCB1, LCB2a, SPTssa, and ORM1, in an inhibited state. A ceramide molecule is sandwiched between ORM1 and LCB2a in the cytosolic membrane leaflet. Ceramide binding is critical for the ORM1-dependent SPT repression, and dihydroceramides and phytoceramides differentially affect this repression. A hybrid β sheet, formed by the amino termini of ORM1 and LCB2a and induced by ceramide binding, stabilizes the amino terminus of ORM1 in an inhibitory conformation. Our findings provide mechanistic insights into sphingolipid homeostatic regulation via the binding of ceramide to the SPT-ORM/ORMDL complex that may have implications for plant-specific processes such as the hypersensitive response for microbial pathogen resistance.
- Department of Chemical Biology, Department of Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China.
Organizational Affiliation: