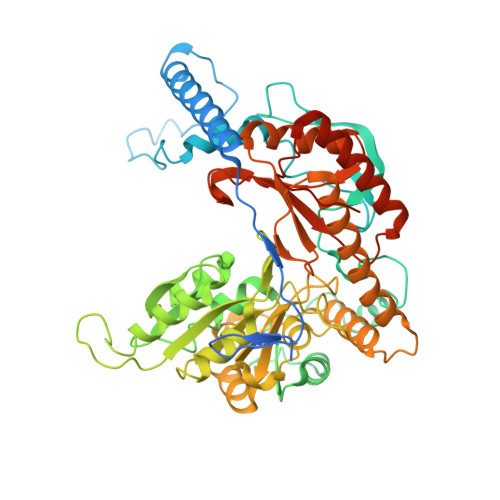
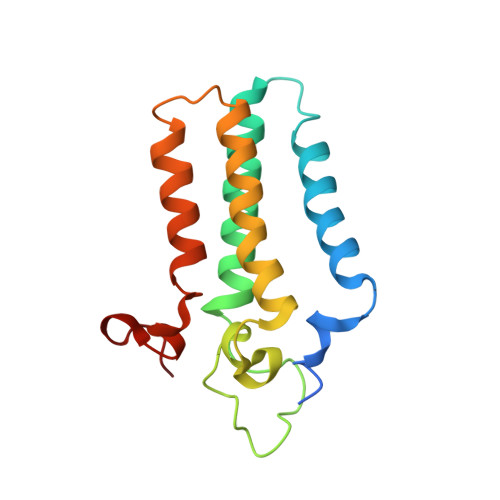
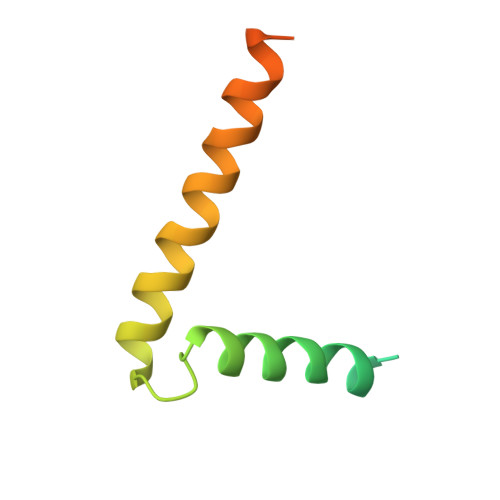
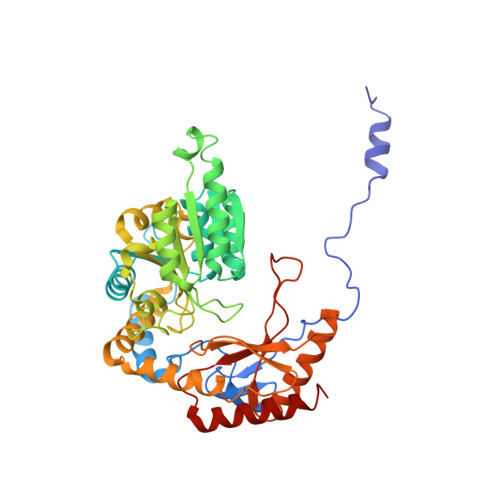
Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Xie, T., Liu, P., Wu, X., Dong, F., Zhang, Z., Yue, J., Mahawar, U., Farooq, F., Vohra, H., Fang, Q., Liu, W., Wattenberg, B.W., Gong, X.(2023) Nat Commun 14: 3475-3475
- PubMed: 37308477
- DOI: https://doi.org/10.1038/s41467-023-39274-y
- Primary Citation of Related Structures:
7YIU, 7YIY, 7YJ1, 7YJ2 - PubMed Abstract:
The ORM/ORMDL family proteins function as regulatory subunits of the serine palmitoyltransferase (SPT) complex, which is the initiating and rate-limiting enzyme in sphingolipid biosynthesis. This complex is tightly regulated by cellular sphingolipid levels, but the sphingolipid sensing mechanism is unknown. Here we show that purified human SPT-ORMDL complexes are inhibited by the central sphingolipid metabolite ceramide. We have solved the cryo-EM structure of the SPT-ORMDL3 complex in a ceramide-bound state. Structure-guided mutational analyses reveal the essential function of this ceramide binding site for the suppression of SPT activity. Structural studies indicate that ceramide can induce and lock the N-terminus of ORMDL3 into an inhibitory conformation. Furthermore, we demonstrate that childhood amyotrophic lateral sclerosis (ALS) variants in the SPTLC1 subunit cause impaired ceramide sensing in the SPT-ORMDL3 mutants. Our work elucidates the molecular basis of ceramide sensing by the SPT-ORMDL complex for establishing sphingolipid homeostasis and indicates an important role of impaired ceramide sensing in disease development.
- Department of Chemical Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, Guangdong, 518055, China.
Organizational Affiliation: