The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii
Wang, W.W., Yi, G.S., Zhou, H., Zhao, Y.X., Wang, Q.S., He, J.H., Yu, F., Xiao, X., Liu, X.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| nuclease RecJ2 | 432 | Methanocaldococcus jannaschii DSM 2661 | Mutation(s): 0 Gene Names: MJ0831 EC: 3.1 | 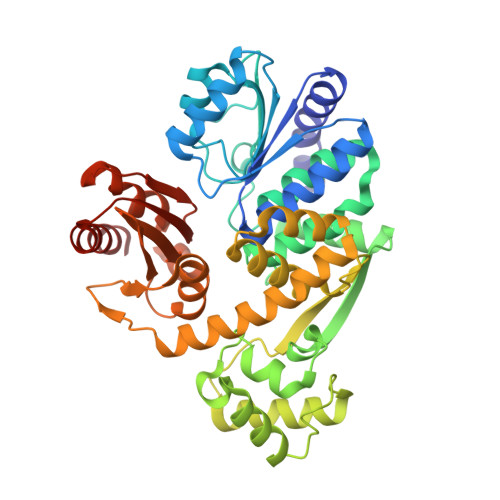 | |
UniProt | |||||
Find proteins for Q58241 (Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)) Explore Q58241 Go to UniProtKB: Q58241 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q58241 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 85.522 | α = 90 |
| b = 177.818 | β = 90 |
| c = 66.297 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32170097 |
| National Natural Science Foundation of China (NSFC) | China | U1832161 |
| National Natural Science Foundation of China (NSFC) | China | 41921006 |