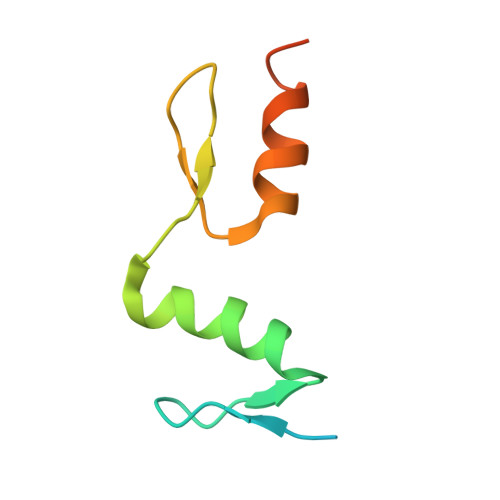
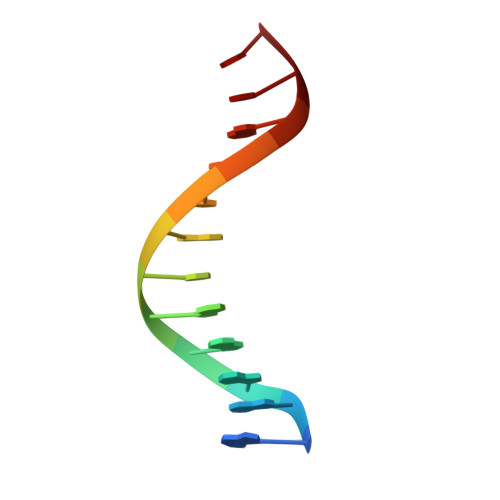
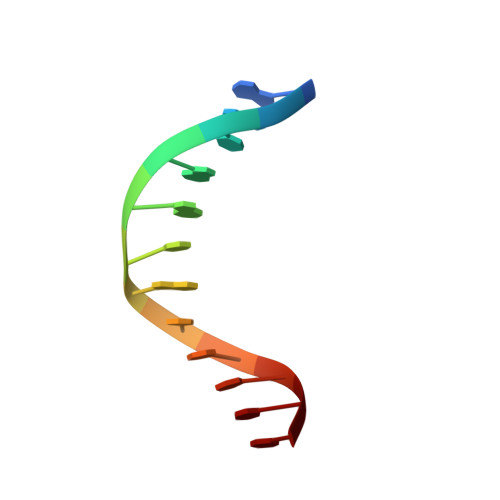
Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
Ru, W., Koga, T., Wang, X., Guo, Q., Gearhart, M.D., Zhao, S., Murphy, M., Kawakami, H., Corcoran, D., Zhang, J., Zhu, Z., Yao, X., Kawakami, Y., Xu, C.(2022) J Biological Chem 298: 102607-102607
- PubMed: 36257403
- DOI: https://doi.org/10.1016/j.jbc.2022.102607
- Primary Citation of Related Structures:
7Y3I, 7Y3K, 7Y3L, 7Y3M - PubMed Abstract:
The Spalt-like 4 transcription factor (SALL4) plays an essential role in controlling the pluripotent property of embryonic stem cells via binding to AT-rich regions of genomic DNA, but structural details on this binding interaction have not been fully characterized. Here, we present crystal structures of the zinc finger cluster 4 (ZFC4) domain of SALL4 (SALL4 ZFC4 ) bound with different dsDNAs containing a conserved AT-rich motif. In the structures, two zinc fingers of SALL4 ZFC4 recognize an AATA tetranucleotide. We also solved the DNA-bound structures of SALL3 ZFC4 and SALL4 ZFC1 . These structures illuminate a common preference for the AATA tetranucleotide shared by ZFC4 of SALL1, SALL3, and SALL4. Furthermore, our cell biology experiments demonstrate that the DNA-binding activity is essential for SALL4 function as DNA-binding defective mutants of mouse Sall4 failed to repress aberrant gene expression in Sall4-/- mESCs. Thus, these analyses provide new insights into the mechanisms of action underlying SALL family proteins in controlling cell fate via preferential targeting to AT-rich sites within genomic DNA during cell differentiation.
- MOE Key Laboratory for Cellular Dynamics, Hefei National Center for Cross-disciplinary Sciences, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, P. R. China.
Organizational Affiliation: