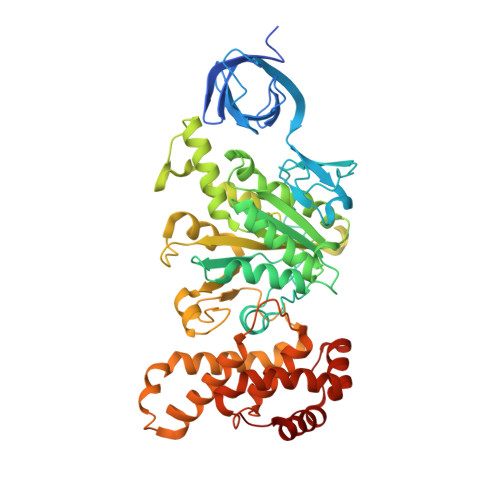
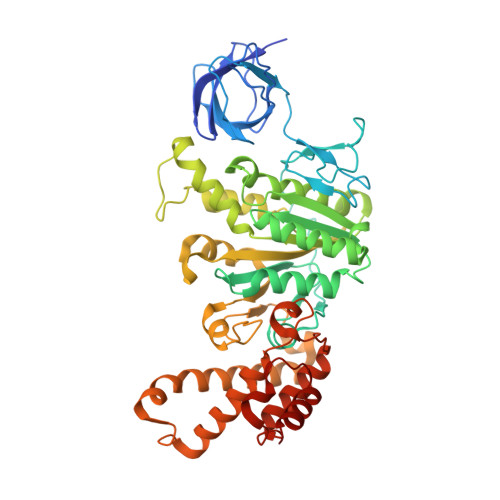
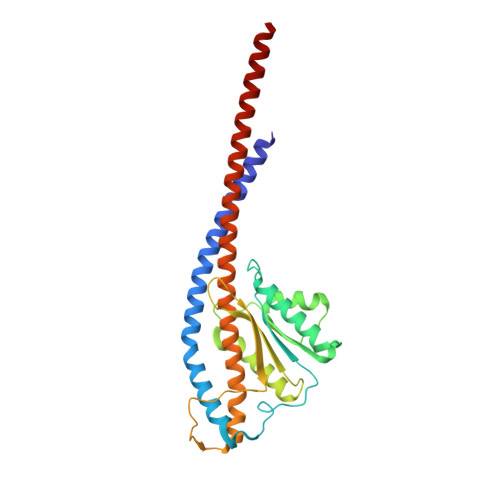
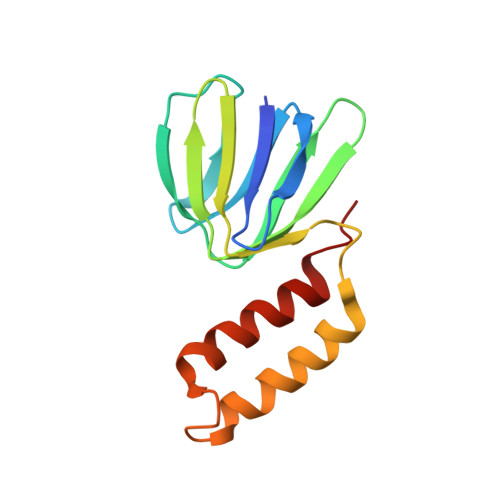
Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Nakano, A., Kishikawa, J.I., Nakanishi, A., Mitsuoka, K., Yokoyama, K.(2022) PNAS Nexus 1: pgac116-pgac116
- PubMed: 36741449
- DOI: https://doi.org/10.1093/pnasnexus/pgac116
- Primary Citation of Related Structures:
7XKH, 7XKO, 7XKP, 7XKQ, 7XKR - PubMed Abstract:
Adenosine triphosphate (ATP) synthases (F 0 F 1 -ATPases) are crucial for all aerobic organisms. F 1 , a water-soluble domain, can catalyze both the synthesis and hydrolysis of ATP with the rotation of the central γε rotor inside a cylinder made of α 3 β 3 in three different conformations (referred to as β E , β TP , and β DP ). In this study, we determined multiple cryo-electron microscopy structures of bacterial F 0 F 1 exposed to different reaction conditions. The structures of nucleotide-depleted F 0 F 1 indicate that the ε subunit directly forces β TP to adopt a closed form independent of the nucleotide binding to β TP . The structure of F 0 F 1 under conditions that permit only a single catalytic β subunit per enzyme to bind ATP is referred to as unisite catalysis and reveals that ATP hydrolysis unexpectedly occurs on β TP instead of β DP , where ATP hydrolysis proceeds in the steady-state catalysis of F 0 F 1 . This indicates that the unisite catalysis of bacterial F 0 F 1 significantly differs from the kinetics of steady-state turnover with continuous rotation of the shaft.
- Department of Molecular Biosciences, Kyoto Sangyo University, Kamigamo-Motoyama, Kita-ku, Kyoto 603-8555, Japan.
Organizational Affiliation: