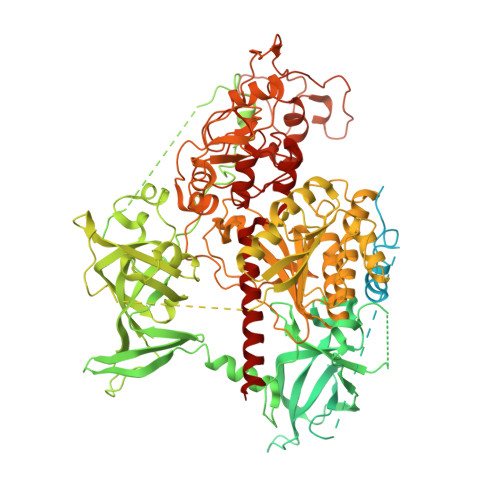
Structural basis for activation of DNMT1.
Kikuchi, A., Onoda, H., Yamaguchi, K., Kori, S., Matsuzawa, S., Chiba, Y., Tanimoto, S., Yoshimi, S., Sato, H., Yamagata, A., Shirouzu, M., Adachi, N., Sharif, J., Koseki, H., Nishiyama, A., Nakanishi, M., Defossez, P.A., Arita, K.(2022) Nat Commun 13: 7130-7130
- PubMed: 36414620
- DOI: https://doi.org/10.1038/s41467-022-34779-4
- Primary Citation of Related Structures:
7XI9, 7XIB - PubMed Abstract:
DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human DNMT1 bound to its two natural activators: hemimethylated DNA and ubiquitinated histone H3. We find that a hitherto unstudied linker, between the RFTS and CXXC domains, plays a key role for activation. It contains a conserved α-helix which engages a crucial "Toggle" pocket, displacing a previously described inhibitory linker, and allowing the DNA Recognition Helix to spring into the active conformation. This is accompanied by large-scale reorganization of the inhibitory RFTS and CXXC domains, allowing the enzyme to gain full activity. Our results therefore provide a mechanistic basis for the activation of DNMT1, with consequences for basic research and drug design.
- Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan.
Organizational Affiliation: