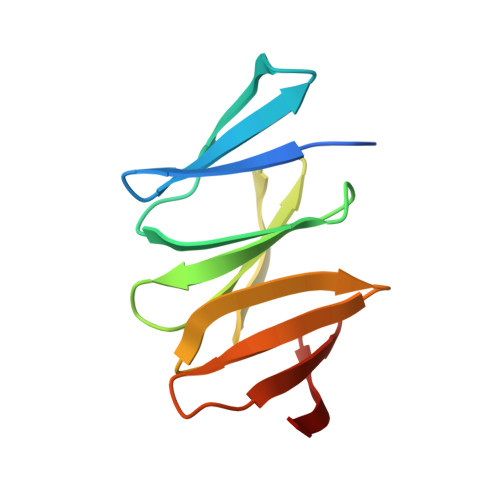
Structural analysis of the virulence gene protein IceA2 from Helicobacter pylori.
Cho, H.Y., Cho, H., Song, W.S., Yoon, S.I.(2022) Biochem Biophys Res Commun 612: 162-168
- PubMed: 35526497
- DOI: https://doi.org/10.1016/j.bbrc.2022.04.090
- Primary Citation of Related Structures:
7XFP - PubMed Abstract:
Helicobacter pylori is a pathogenic bacterium that causes gastric ulcers and cancer. Among the diverse virulence genes of H. pylori, the IceA gene was identified to be expressed upon adherence to host cells. The IceA gene has two alleles, iceA1 and iceA2, which encode completely different proteins. IceA1 protein was shown to exert endonuclease activity, whereas IceA2 has never been analyzed at the molecular level. Based on a sequence analysis, IceA2 proteins differ in length depending on the strain and are classified into five groups (A-E). To structurally characterize IceA2, we determined the crystal structure of group-D IceA2 (IceA2 sD ) and performed a modeling-based comparative analysis of IceA2 groups. IceA2 sD consists of three β-sheet repeats and serially arranges them like the β-propeller structure of the WD40 domain. However, each β-sheet of IceA2 is stabilized using a unique structural motif that is not observed in WD40. Moreover, IceA2 sD lacks an additionally appended β-strand and does not form the Velcro-like closure of WD40. Therefore, IceA2 sD adopts a curved rod-like structure rather than an enclosed circular structure in WD40. IceA2 proteins contain 1-4 β-sheet modules depending on the groups and are modeled to be highly diverse in size and shape.
- Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, Republic of Korea.
Organizational Affiliation: