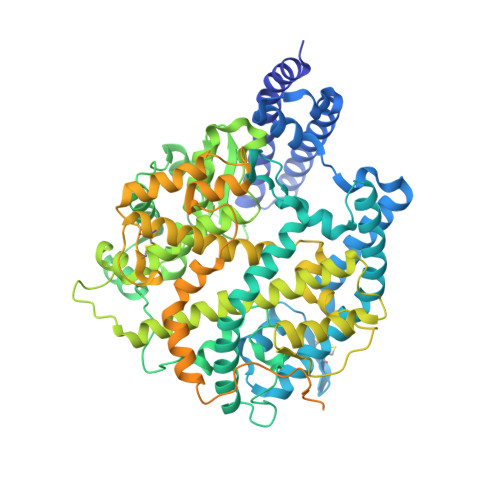
Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Xu, Z., Kang, X., Han, P., Du, P., Li, L., Zheng, A., Deng, C., Qi, J., Zhao, X., Wang, Q., Liu, K., Gao, G.F.(2022) Nat Commun 13: 3547-3547
- PubMed: 35729237
- DOI: https://doi.org/10.1038/s41467-022-31276-6
- Primary Citation of Related Structures:
7XBY - PubMed Abstract:
The origin and host range of SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19), are important scientific questions as they might provide insight into understanding of the potential future spillover to infect humans. Here, we tested the binding between equine angiotensin converting enzyme 2 (eqACE2) and the receptor binding domains (RBDs) of SARS-CoV, SARS-CoV-2 prototype (PT) and variant of concerns (VOCs), as well as their close relatives bat-origin coronavirus (CoV) RaTG13 and pangolin-origin CoVs GX/P2V/2017 and GD/1/2019. We also determined the crystal structures of eqACE2/RaTG13-RBD, eqACE2/SARS-CoV-2 PT-RBD and eqACE2/Omicron BA.1-RBD. We identified S494 of SARS-COV-2 PT-RBD as an important residue in the eqACE2/SARS-COV-2 PT-RBD interaction and found that N501Y, the commonly recognized enhancing mutation, attenuated the binding affinity with eqACE2. Our work demonstrates that horses are potential targets for SARS-CoV-2 and highlights the importance of continuous surveillance on SARS-CoV-2 and related CoVs to prevent spillover events.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China.
Organizational Affiliation: