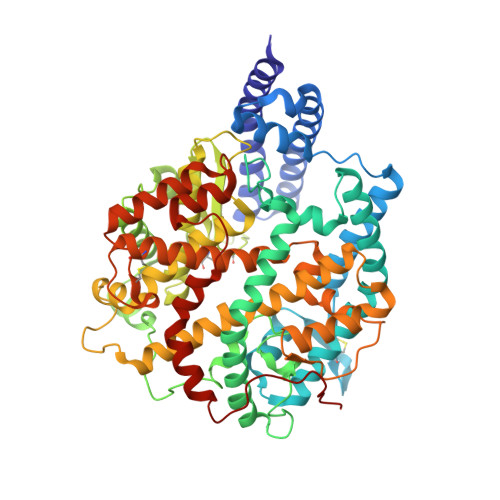
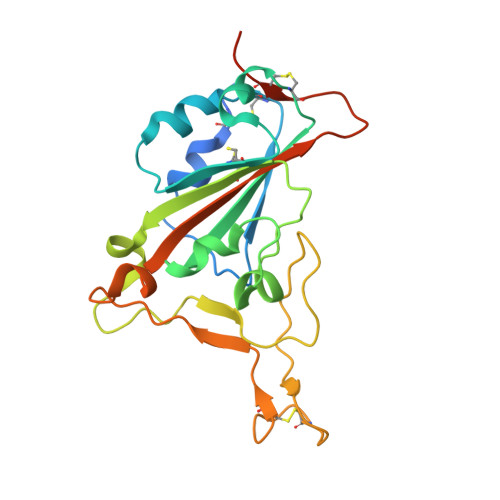
Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Tang, L., Zhang, D., Han, P., Kang, X., Zheng, A., Xu, Z., Zhao, X., Wang, V.Y., Qi, J., Wang, Q., Liu, K., Gao, G.F.(2022) Int J Biol Sci 18: 4658-4668
- PubMed: 35874946
- DOI: https://doi.org/10.7150/ijbs.73640
- Primary Citation of Related Structures:
7XA7 - PubMed Abstract:
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a global pandemic. Intermediate horseshoe bats ( Rhinolophus affinis ) are hosts of RaTG13, the second most phylogenetically related viruses to SARS-CoV-2. We report the binding between intermediate horseshoe bat ACE2 (bACE2-Ra) and SARS-CoV-2 receptor-binding domain (RBD), supporting the pseudotyped SARS-CoV-2 viral infection. A 3.3 Å resolution crystal structure of the bACE2-Ra/SARS-CoV-2 RBD complex was determined. The interaction networks of Patch 1 showed differences in R34 and E35 of bACE2-Ra compared to hACE2 and big-eared horseshoe bat ACE2 (bACE2-Rm). The E35K substitution, existing in other species, significantly enhanced the binding affinity owing to its electrostatic attraction with E484 of SARS-CoV-2 RBD. Furthermore, bACE2-Ra showed extensive support for the SARS-CoV-2 variants. These results broaden our knowledge of the ACE2/RBD interaction mechanism and emphasize the importance of continued surveillance of intermediate horseshoe bats to prevent spillover risk.
- CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: