Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Qian, Y., Ma, Z., Liu, C., Li, X., Zhu, X., Wang, N., Xu, Z., Xia, R., Liang, J., Duan, Y., Yin, H., Xiong, Y., Zhang, A., Guo, C., Chen, Z., Huang, Z., He, Y.(2022) Mol Cell 82: 4340-4352.e6
- PubMed: 36309016
- DOI: https://doi.org/10.1016/j.molcel.2022.10.009
- Primary Citation of Related Structures:
7WY5, 7WY8, 7WYB, 7X10 - PubMed Abstract:
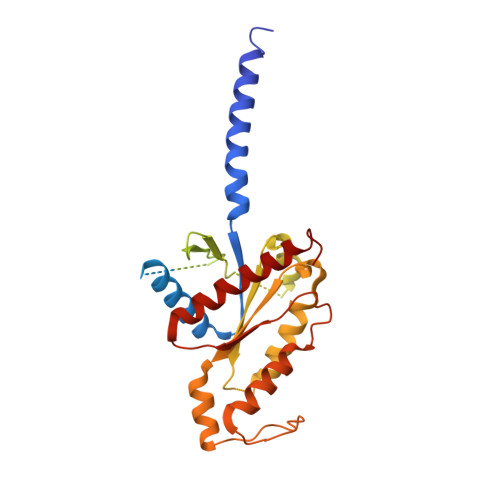
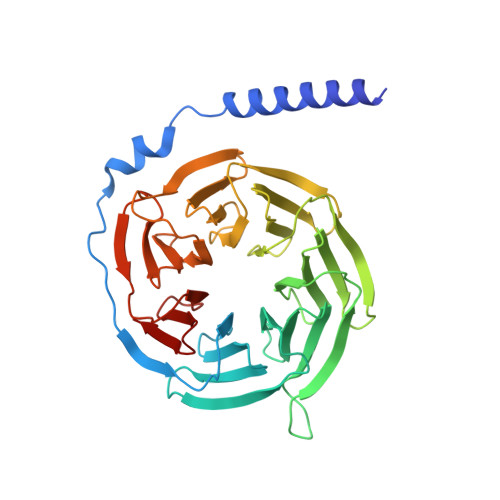
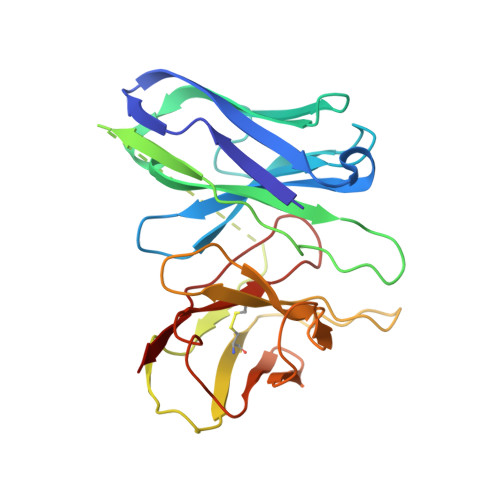
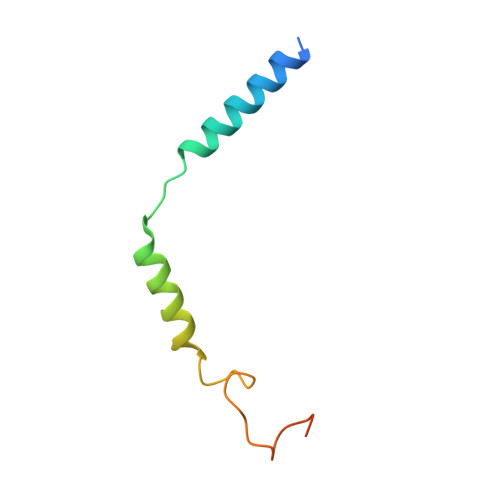
Adhesion G-protein-coupled receptors (aGPCRs) play key roles in a diversity of physiologies. A hallmark of aGPCR activation is the removal of the inhibitory GAIN domain and the dipping of the cleaved stalk peptide into the ligand-binding pocket of receptors; however, the detailed mechanism remains obscure. Here, we present cryoelectron microscopy (cryo-EM) structures of ADGRL3 in complex with G q , G s , G i , and G 12 . The structures reveal unique ligand-engaging mode, distinctive activation conformation, and key mechanisms of aGPCR activation. The structures also reveal the uncharted structural information of GPCR/G 12 coupling. A comparison of G q , G s , G i , and G 12 engagements with ADGRL3 reveals the key determinant of G-protein coupling on the far end of αH5 of Gα. A detailed analysis of the engagements allows us to design mutations that specifically enhance one pathway over others. Taken together, our study lays the groundwork for understanding aGPCR activation and G-protein-coupling selectivity.
- Laboratory of Receptor Structure and Signaling, HIT Center for Life Sciences, Harbin Institute of Technology, Harbin 150001, China; HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150080, China.
Organizational Affiliation: