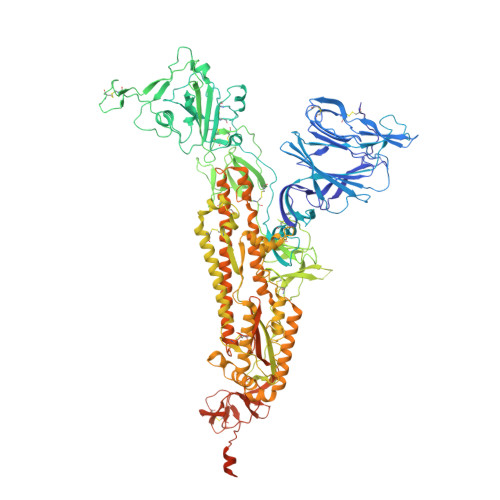
Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Zhao, M.M., Zhu, Y., Zhang, L., Zhong, G., Tai, L., Liu, S., Yin, G., Lu, J., He, Q., Li, M.J., Zhao, R.X., Wang, H., Huang, W., Fan, C., Shuai, L., Wen, Z., Wang, C., He, X., Chen, Q., Liu, B., Xiong, X., Bu, Z., Wang, Y., Sun, F., Yang, J.K.(2022) Cell Discov 8: 53