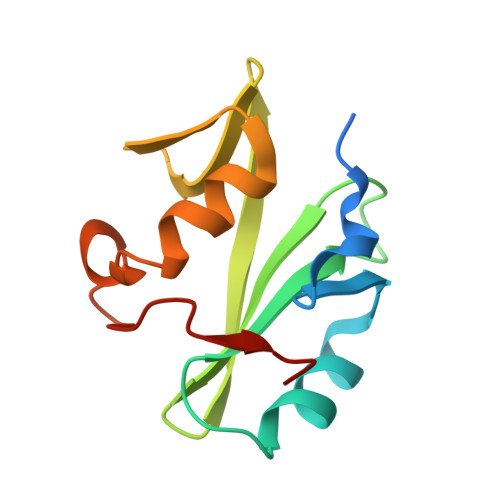
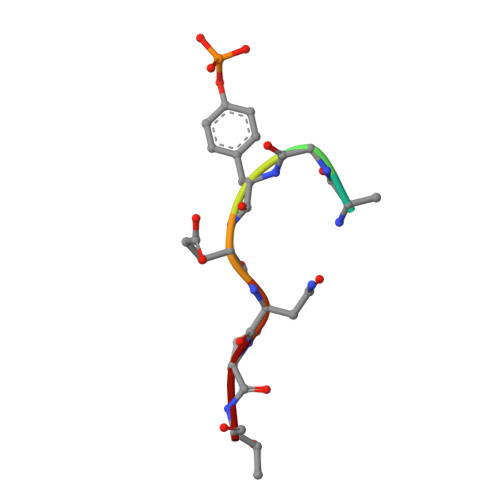
Vav2 is a novel APP-interacting protein that regulates APP protein level.
Zhang, Y., Yang, X., Liu, Y., Ge, L., Wang, J., Sun, X., Wu, B., Wang, J.(2022) Sci Rep 12: 12752-12752
- PubMed: 35882892
- DOI: https://doi.org/10.1038/s41598-022-16883-z
- Primary Citation of Related Structures:
7WFY - PubMed Abstract:
Amyloid precursor protein (APP) is a transmembrane protein that plays critical role in the pathogenesis of Alzheimer's disease (AD). It is also involved in many types of cancers. Increasing evidence has shown that the tyrosine phosphorylation site Y682 in the intracellular tail of APP is crucial for APP function. Here, we report that Vav2, a guanine nucleotide exchange factor (GEF) for Rho family GTPase, is a novel interaction partner of APP. We found that Vav2-SH2 domain was able to bind directly to the Y682-phosphorylated intracellular tail of APP through isothermal titration calorimetry and NMR titrating experiments. The crystal structure of Vav2-SH2 in complex with an APP-derived phosphopeptide was determined to understand the structural basis of this recognition specificity. The interaction of APP and Vav2 in a full-length manner was further confirmed in cells by GST pull-down, co-immunoprecipitation and immunofluorescence staining experiments. In addition, we found overexpression of Vav2 could inhibit APP degradation and markedly increase the protein levels of APP and its cleavage productions in 20E2 cells, and this function of Vav2 required a functional SH2 domain.
- High Magnetic Field Laboratory, Key Laboratory of High Magnetic Field and Ion Beam Physical Biology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, Anhui, China.
Organizational Affiliation: