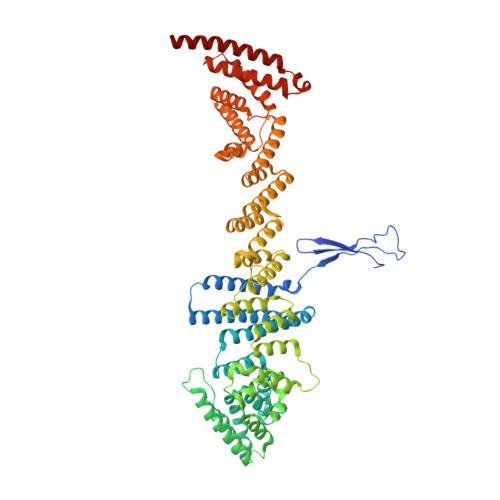
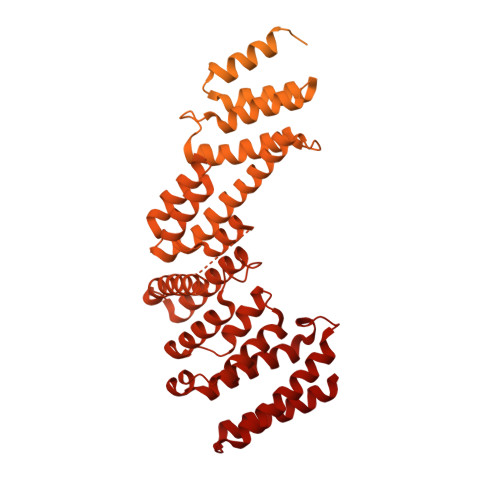
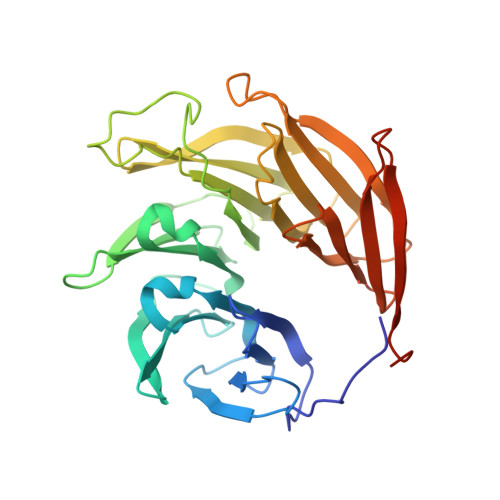
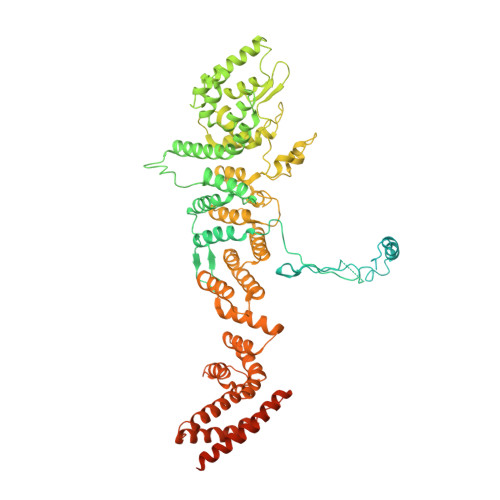
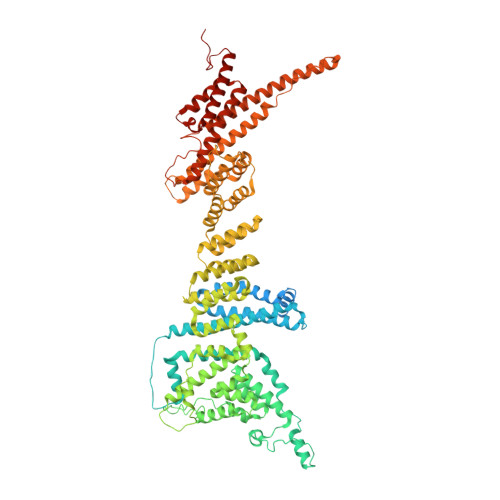
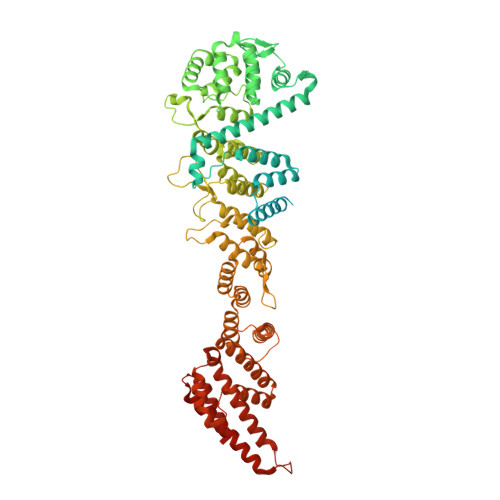
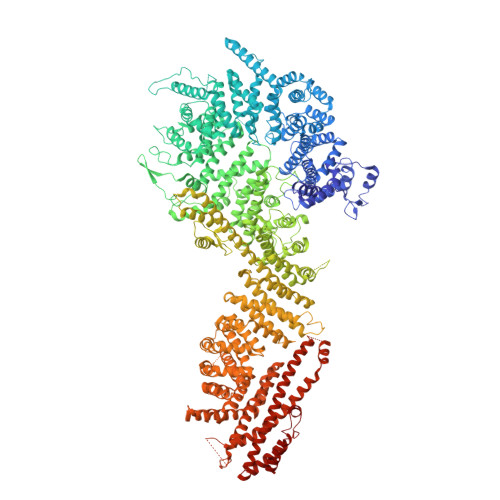
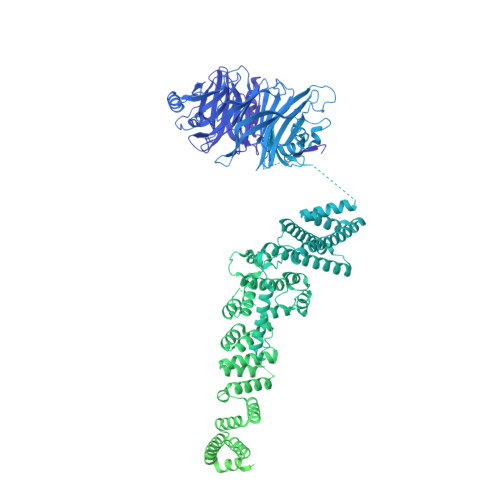
Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Huang, G., Zhan, X., Zeng, C., Zhu, X., Liang, K., Zhao, Y., Wang, P., Wang, Q., Zhou, Q., Tao, Q., Liu, M., Lei, J., Yan, C., Shi, Y.(2022) Cell Res 32: 349-358
- PubMed: 35177819
- DOI: https://doi.org/10.1038/s41422-021-00610-w
- Primary Citation of Related Structures:
7WB4 - PubMed Abstract:
Nuclear pore complex (NPC) shuttles cargo across the nuclear envelope. Here we present single-particle cryo-EM structure of the nuclear ring (NR) subunit from Xenopus laevis NPC at an average resolution of 5.6 Å. The NR subunit comprises two 10-membered Y complexes, each with the nucleoporin ELYS closely associating with Nup160 and Nup37 of the long arm. Unlike the cytoplasmic ring (CR) or inner ring (IR), the NR subunit contains only one molecule each of Nup205 and Nup93. Nup205 binds both arms of the Y complexes and interacts with the stem of inner Y complex from the neighboring subunit. Nup93 connects the stems of inner and outer Y complexes within the same NR subunit, and places its N-terminal extended helix into the axial groove of Nup205 from the neighboring subunit. Together with other structural information, we have generated a composite atomic model of the central ring scaffold that includes the NR, IR, and CR. The IR is connected to the two outer rings mainly through Nup155. This model facilitates functional understanding of vertebrate NPC.
- Westlake Laboratory of Life Sciences and Biomedicine, 18 Shilongshan Road, Hangzhou, Zhejiang, China. huanggaoxingyu@westlake.edu.cn.
Organizational Affiliation: