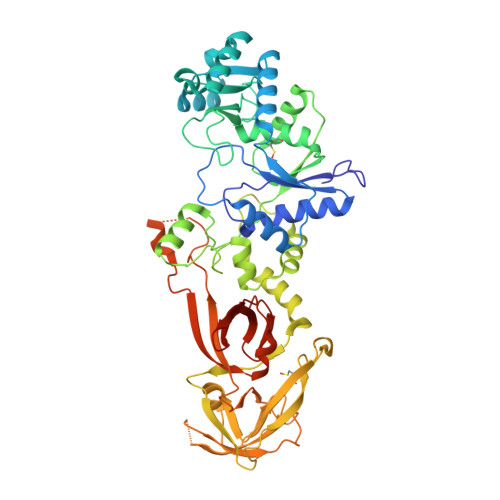
Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Sharma, V.K., Chhibber-Goel, J., Yogavel, M., Sharma, A.(2022) Mol Biochem Parasitol 253: 111530-111530
- PubMed: 36370911
- DOI: https://doi.org/10.1016/j.molbiopara.2022.111530
- Primary Citation of Related Structures:
7WAI, 7WAJ, 7WAK, 7WAL, 7WAO - PubMed Abstract:
Aminoacyl-tRNA synthetases (aaRSs) are essential enzymes in protein translation machinery that provide the charged tRNAs needed for protein synthesis. Over the past decades, aaRSs have been studied as anti-parasitic, anti-bacterial, and anti-fungal drug targets. This study focused on the cytoplasmic glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum, which belongs to class Ib in aaRSs. GluRS unlike most other aaRSs requires tRNA to activate its cognate amino acid substrate L-Glutamate (L-Glu), and fails to form an intermediate adenylate complex in the absence of tRNA. The crystal structures of the Apo, ATP, and ADP-bound forms of Plasmodium falciparum glutamyl-tRNA synthetase (PfGluRS) were solved at 2.1 Å, 2.2 Å, and 2.8 Å respectively. The structural comparison of the Apo- and ATP-bound holo-forms of PfGluRS showed considerable conformational changes in the loop regions around the ATP-binding pocket of the enzyme. Biophysical characterization of the PfGluRS showed binding of the enzyme substrates L-Gluand ATP.. The sequence and structural conservation were evident across GluRS compared to other species. The structural dissection of the PfGluRS gives insight into the critical residues involved in the binding of ATP substrate, which can be harvested to develop new antimalarial drugs.
- Molecular Medicine - Structural Parasitology, International Centre for Genetic Engineering and Biotechnology, New Delhi 110067, India.
Organizational Affiliation: