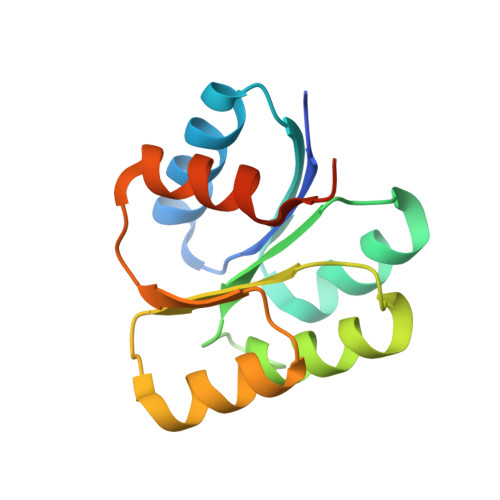
REC domain stabilizes the active heptamer of sigma 54 -dependent transcription factor, FleR from Pseudomonas aeruginosa.
Sahoo, P.K., Jain, D.(2023) iScience 26: 108397-108397
- PubMed: 38058307
- DOI: https://doi.org/10.1016/j.isci.2023.108397
- Primary Citation of Related Structures:
7W9H - PubMed Abstract:
Motility in Pseudomonas aeruginosa is mediated through a single, polar flagellum, which is essential for virulence, colonization, and biofilm formation. FleSR, a two-component system (TCS), serves as a critical checkpoint in flagellar assembly. FleR is a σ 54 -dependent response regulator that undergoes phosphorylation via cognate sensor kinase FleS for the assembly of the functionally active form. The active form remodels the σ 54 -RNAP complex to initiate transcription. Small-angle X-ray scattering, crystallography, and negative staining electron microscopy reconstructions of FleR revealed that it exists predominantly as a dimer in the inactive form with low ATPase activity and assembles into heptamers upon phosphorylation with amplified ATPase activity. We establish that receiver (REC) domain stabilizes the heptamers and is indispensable for assembly of the functional phosphorylated form of FleR. The structural, biochemical, and in vivo complementation assays provide details of the phosphorylation-mediated assembly of FleR to regulate the expression of flagellar genes.
- Transcription Regulation Lab, Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad 121001, India.
Organizational Affiliation: