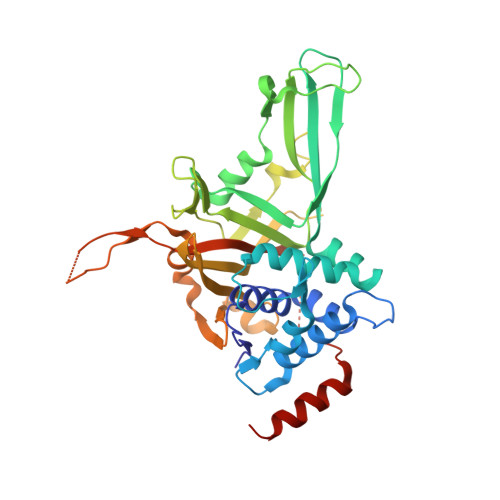
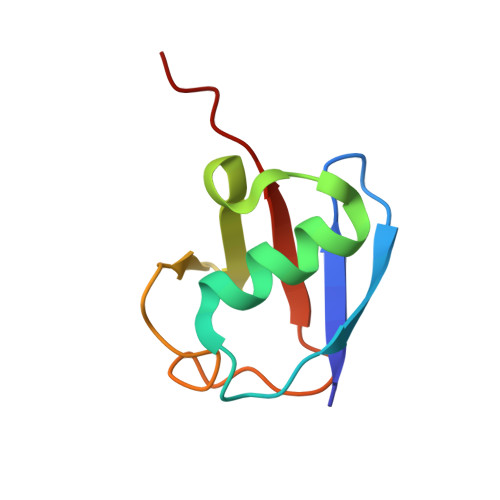
Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
Xu, G., Su, H., Lu, L., Liu, X., Zhao, L., Tang, B., Ming, Z.(2022) J Mol Biology 434: 167634-167634
- PubMed: 35588869
- DOI: https://doi.org/10.1016/j.jmb.2022.167634
- Primary Citation of Related Structures:
7W3R, 7W3U - PubMed Abstract:
Ubiquitination, an important posttranslational modification, participates in virtually all aspects of cellular functions and is reversed by deubiquitinating enzymes (DUBs). Ubiquitin-specific protease 34 (USP34) plays an essential role in cancer, neurodegenerative diseases, and osteogenesis. Despite its functional importance, how USP34 recognizes ubiquitin and catalyzes deubiquitination remains structurally uncharacterized. Here, we report the crystal structures of the USP34 catalytic domain in free state and after binding with ubiquitin. In the free state, USP34 adopts an inactive conformation, which contains a misaligned catalytic histidine in the triad. Comparison of USP34 structures before and after ubiquitin binding reveals a structural basis for ubiquitin recognition and elucidates a mechanism by which the catalytic triad is realigned. Transition from an open inactive state to a relatively closed active state is coupled to a process by which the "fingertips" of USP34 intimately grip ubiquitin, and this has not been reported before. Our structural and biochemical analyses provide important insights into the catalytic mechanism and ubiquitin recognition of USP34.
- State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, PR China.
Organizational Affiliation: