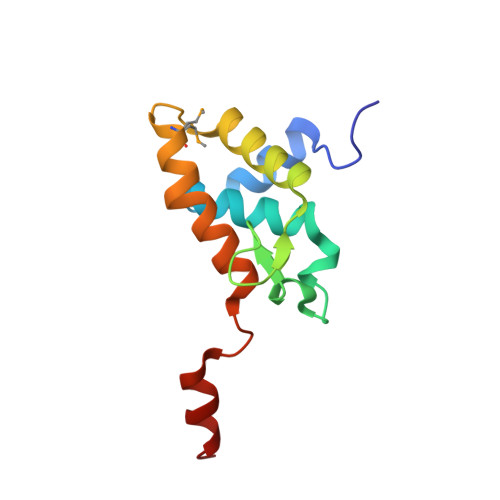
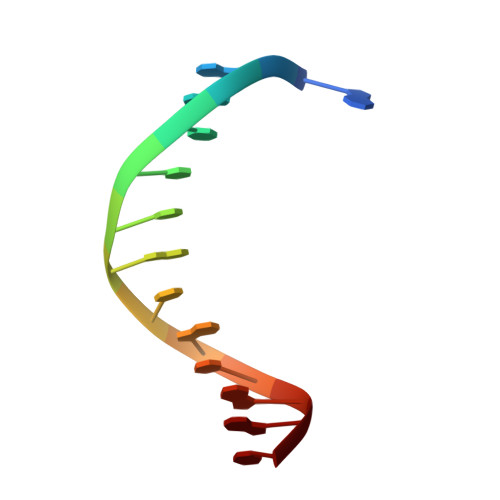
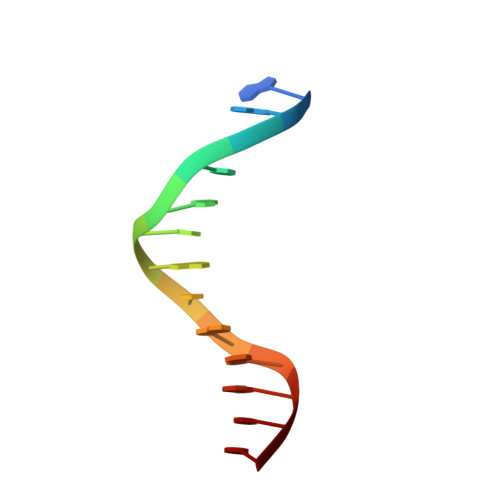
Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Zheng, L., Liu, J., Niu, L., Kamran, M., Yang, A.W.H., Jolma, A., Dai, Q., Hughes, T.R., Patel, D.J., Zhang, L., Prasanth, S.G., Yu, Y., Ren, A., Lai, E.C.(2022) Genes Dev 36: 225-240
- PubMed: 35144965
- DOI: https://doi.org/10.1101/gad.348993.121
- Primary Citation of Related Structures:
7W27 - PubMed Abstract:
The BEN domain is a recently recognized DNA binding module that is present in diverse metazoans and certain viruses. Several BEN domain factors are known as transcriptional repressors, but, overall, relatively little is known of how BEN factors identify their targets in humans. In particular, X-ray structures of BEN domain:DNA complexes are only known for Drosophila factors bearing a single BEN domain, which lack direct vertebrate orthologs. Here, we characterize several mammalian BEN domain (BD) factors, including from two NACC family BTB-BEN proteins and from BEND3, which has four BDs. In vitro selection data revealed sequence-specific binding activities of isolated BEN domains from all of these factors. We conducted detailed functional, genomic, and structural studies of BEND3. We show that BD4 is a major determinant for in vivo association and repression of endogenous BEND3 targets. We obtained a high-resolution structure of BEND3-BD4 bound to its preferred binding site, which reveals how BEND3 identifies cognate DNA targets and shows differences with one of its non-DNA-binding BEN domains (BD1). Finally, comparison with our previous invertebrate BEN structures, along with additional structural predictions using AlphaFold2 and RoseTTAFold, reveal distinct strategies for target DNA recognition by different types of BEN domain proteins. Together, these studies expand the DNA recognition activities of BEN factors and provide structural insights into sequence-specific DNA binding by mammalian BEN proteins.
- The Eighth Affiliated Hospital, Sun Yat-Sen University, Shenzhen, Guangdong 518033, China.
Organizational Affiliation: