ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Liu, D., Dredge, B.K., Bert, A.G., Pillman, K.A., Toubia, J., Guo, W., Dyakov, B.J.A., Migault, M.M., Conn, V.M., Conn, S.J., Gregory, P.A., Gingras, A.C., Patel, D., Wu, B., Goodall, G.J.(2023) Nucleic Acids Res
- PubMed: 38015468
- DOI: https://doi.org/10.1093/nar/gkad1138
- Primary Citation of Related Structures:
7VKI - PubMed Abstract:
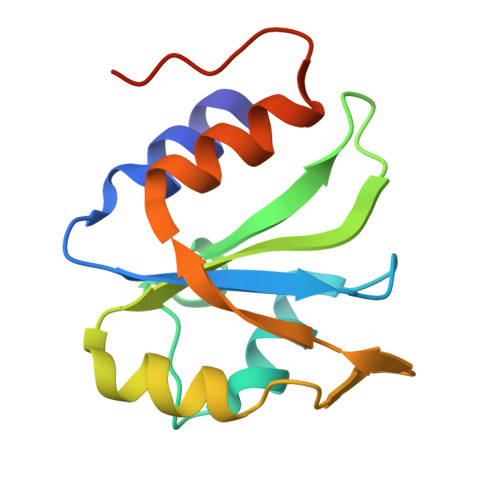
While the majority of circRNAs are formed from infrequent back-splicing of exons from protein coding genes, some can be produced at quite high level and in a regulated manner. We describe the regulation, biogenesis and function of circDOCK1(2-27), a large, abundant circular RNA that is highly regulated during epithelial-mesenchymal transition (EMT) and whose formation depends on the epithelial splicing regulator ESRP1. CircDOCK1(2-27) synthesis in epithelial cells represses cell motility both by diverting transcripts from DOCK1 mRNA production to circRNA formation and by direct inhibition of migration by the circRNA. HITS-CLIP analysis and CRISPR-mediated deletions indicate ESRP1 controls circDOCK1(2-27) biosynthesis by binding a GGU-containing repeat region in intron 1 and detaining its splicing until Pol II completes its 157 kb journey to exon 27. Proximity-dependent biotinylation (BioID) assay suggests ESRP1 may modify the RNP landscape of intron 1 in a way that disfavours communication of exon 1 with exon 2, rather than physically bridging exon 2 to exon 27. The X-ray crystal structure of RNA-bound ESRP1 qRRM2 domain reveals it binds to GGU motifs, with the guanines embedded in clamp-like aromatic pockets in the protein.
- Centre for Cancer Biology, SA Pathology and University of South Australia, Adelaide, SA 5000, Australia.
Organizational Affiliation: