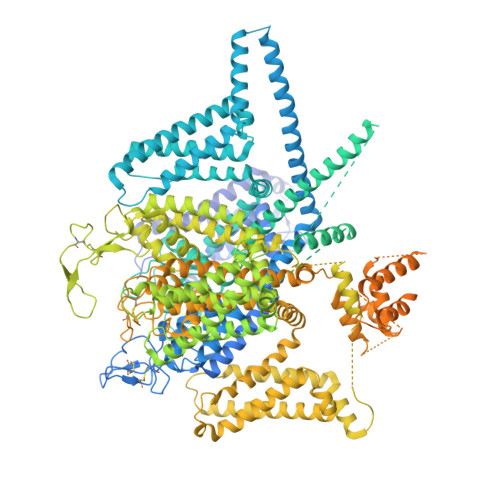
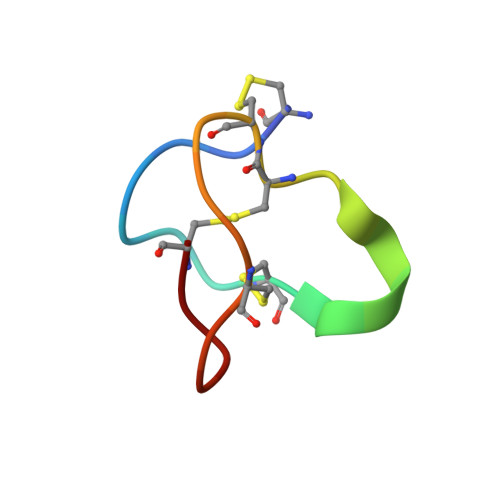
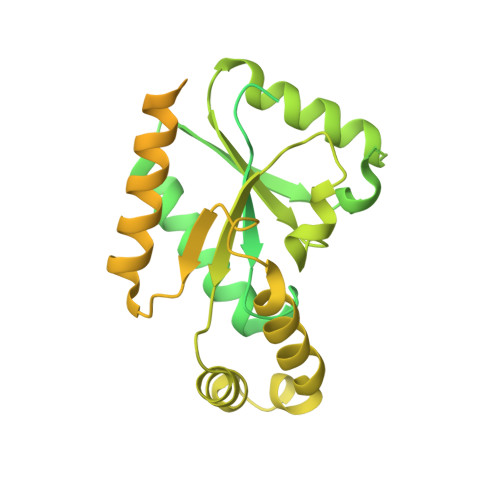
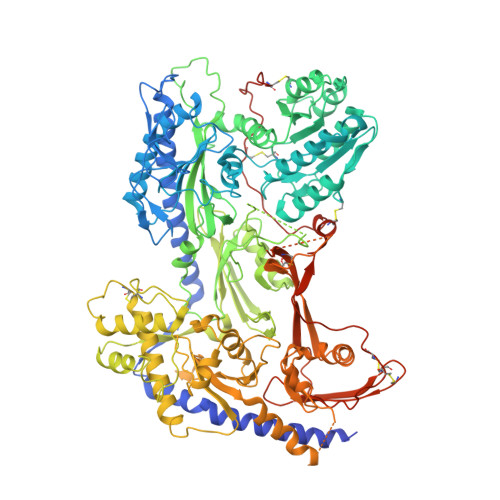
Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Dong, Y., Gao, Y., Xu, S., Wang, Y., Yu, Z., Li, Y., Li, B., Yuan, T., Yang, B., Zhang, X.C., Jiang, D., Huang, Z., Zhao, Y.(2021) Cell Rep 37: 109931-109931
- PubMed: 34731621
- DOI: https://doi.org/10.1016/j.celrep.2021.109931
- Primary Citation of Related Structures:
7VFS, 7VFU, 7VFV, 7VFW - PubMed Abstract:
N-type voltage-gated calcium (Ca V ) channels mediate Ca 2+ influx at presynaptic terminals in response to action potentials and play vital roles in synaptogenesis, release of neurotransmitters, and nociceptive transmission. Here, we elucidate a cryo-electron microscopy (cryo-EM) structure of the human Ca V 2.2 complex in apo, ziconotide-bound, and two Ca V 2.2-specific pore blockers-bound states. The second voltage-sensing domain (VSD) is captured in a resting-state conformation, trapped by a phosphatidylinositol 4,5-bisphosphate (PIP 2 ) molecule, which is distinct from the other three VSDs of Ca V 2.2, as well as activated VSDs observed in previous structures of Ca V channels. This structure reveals the molecular basis for the unique inactivation process of Ca V 2.2 channels, in which the intracellular gate formed by S6 helices is closed and a W-helix from the domain II-III linker stabilizes closed-state inactivation. The structures of this inactivated, drug-bound complex lay a solid foundation for developing new state-dependent blockers for treatment of chronic pain.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; State Key Laboratory of Brain and Cognitive Science, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Organizational Affiliation: