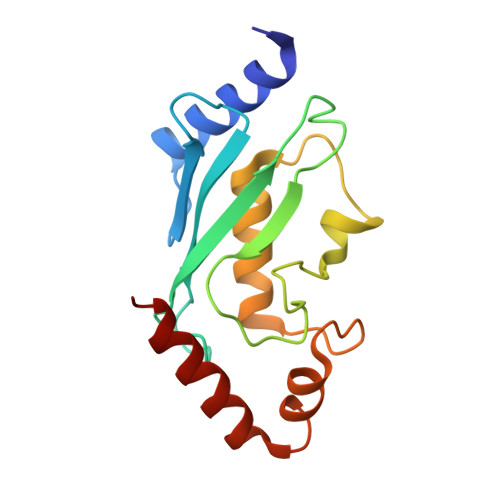
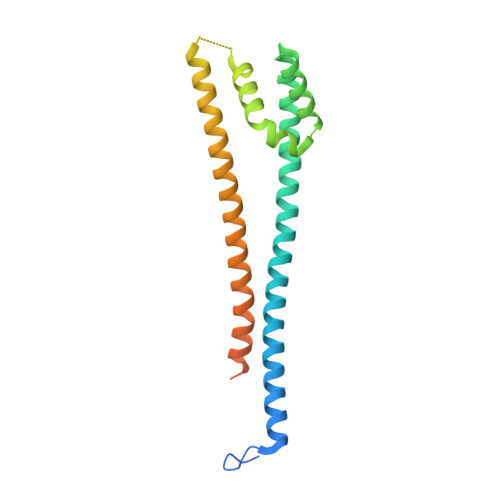
Structure and functional determinants of Rad6-Bre1 subunits in the histone H2B ubiquitin-conjugating complex.
Shukla, P.K., Bissell, J.E., Kumar, S., Pokhrel, S., Palani, S., Radmall, K.S., Obidi, O., Parnell, T.J., Brasch, J., Shrieve, D.C., Chandrasekharan, M.B.(2023) Nucleic Acids Res 51: 2117-2136
- PubMed: 36715322
- DOI: https://doi.org/10.1093/nar/gkad012
- Primary Citation of Related Structures:
7UV8, 7UVC - PubMed Abstract:
The conserved complex of the Rad6 E2 ubiquitin-conjugating enzyme and the Bre1 E3 ubiquitin ligase catalyzes histone H2B monoubiquitination (H2Bub1), which regulates chromatin dynamics during transcription and other nuclear processes. Here, we report a crystal structure of Rad6 and the non-RING domain N-terminal region of Bre1, which shows an asymmetric homodimer of Bre1 contacting a conserved loop on the Rad6 'backside'. This contact is distant from the Rad6 catalytic site and is the location of mutations that impair telomeric silencing in yeast. Mutational analyses validated the importance of this contact for the Rad6-Bre1 interaction, chromatin-binding dynamics, H2Bub1 formation and gene expression. Moreover, the non-RING N-terminal region of Bre1 is sufficient to confer nucleosome binding ability to Rad6 in vitro. Interestingly, Rad6 P43L protein, an interaction interface mutant and equivalent to a cancer mutation in the human homolog, bound Bre1 5-fold more tightly than native Rad6 in vitro, but showed reduced chromatin association of Bre1 and reduced levels of H2Bub1 in vivo. These surprising observations imply conformational transitions of the Rad6-Bre1 complex during its chromatin-associated functional cycle, and reveal the differential effects of specific disease-relevant mutations on the chromatin-bound and unbound states. Overall, our study provides structural insights into Rad6-Bre1 interaction through a novel interface that is important for their biochemical and biological responses.
- Department of Radiation Oncology, University of Utah School of Medicine, Salt Lake City, UT 84112, USA.
Organizational Affiliation: