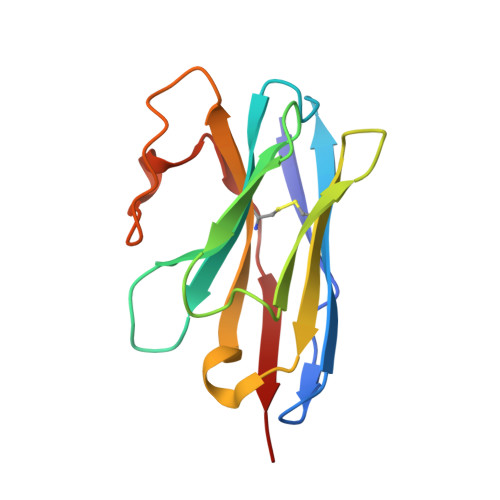
Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Liu, Z., Lee, P.G., Krez, N., Lam, K.H., Liu, H., Przykopanski, A., Chen, P., Yao, G., Zhang, S., Tremblay, J.M., Perry, K., Shoemaker, C.B., Rummel, A., Dong, M., Jin, R.(2023) Nat Commun 14: 2338-2338
- PubMed: 37095076
- DOI: https://doi.org/10.1038/s41467-023-37860-8
- Primary Citation of Related Structures:
7UIA, 7UIB, 7UIE - PubMed Abstract:
Botulinum neurotoxin E (BoNT/E) is one of the major causes of human botulism and paradoxically also a promising therapeutic agent. Here we determined the co-crystal structures of the receptor-binding domain of BoNT/E (H C E) in complex with its neuronal receptor synaptic vesicle glycoprotein 2A (SV2A) and a nanobody that serves as a ganglioside surrogate. These structures reveal that the protein-protein interactions between H C E and SV2 provide the crucial location and specificity information for H C E to recognize SV2A and SV2B, but not the closely related SV2C. At the same time, H C E exploits a separated sialic acid-binding pocket to mediate recognition of an N-glycan of SV2. Structure-based mutagenesis and functional studies demonstrate that both the protein-protein and protein-glycan associations are essential for SV2A-mediated cell entry of BoNT/E and for its potent neurotoxicity. Our studies establish the structural basis to understand the receptor-specificity of BoNT/E and to engineer BoNT/E variants for new clinical applications.
- Department of Physiology and Biophysics, University of California, Irvine, Irvine, CA, 92697, USA.
Organizational Affiliation: