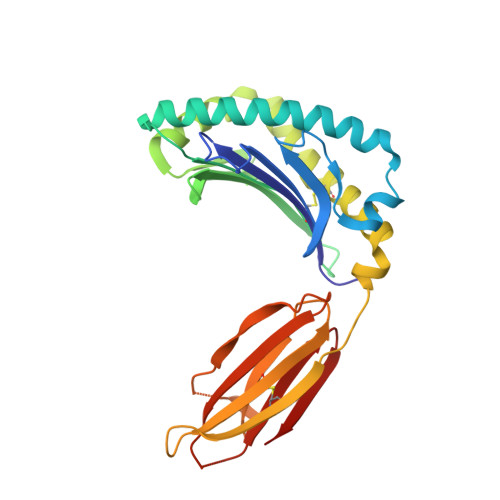
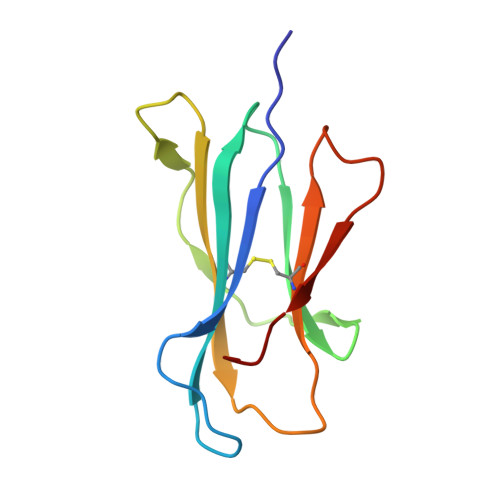
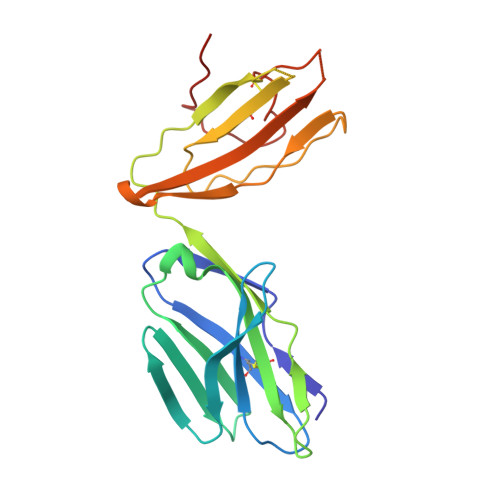
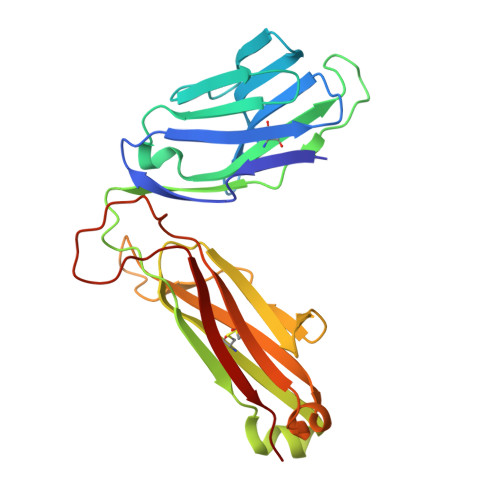
Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
Wang, C.J.H., Awad, W., Liu, L., Mak, J.Y.W., Veerapen, N., Illing, P.T., Purcell, A.W., Eckle, S.B.G., McCluskey, J., Besra, G.S., Fairlie, D.P., Rossjohn, J., Le Nours, J.(2022) J Biological Chem 298: 102714-102714
- PubMed: 36403855
- DOI: https://doi.org/10.1016/j.jbc.2022.102714
- Primary Citation of Related Structures:
7UFJ - PubMed Abstract:
The Major Histocompatibility Complex class I-related protein 1 (MR1) presents small molecule metabolites, drugs, and drug-like molecules that are recognized by MR1-reactive T cells. While we have an understanding of how antigens bind to MR1 and upregulate MR1 cell surface expression, a quantitative, cell-free, assessment of MR1 ligand-binding affinity was lacking. Here, we developed a fluorescence polarization-based assay in which fluorescent MR1 ligand was loaded into MR1 protein in vitro and competitively displaced by candidate ligands over a range of concentrations. Using this assay, ligand affinity for MR1 could be differentiated as strong (IC 50 < 1 μM), moderate (1 μM < IC 50 < 100 μM), and weak (IC 50 > 100 μM). We demonstrated a clear correlation between ligand-binding affinity for MR1, the presence of a covalent bond between MR1 and ligand, and the number of salt bridge and hydrogen bonds formed between MR1 and ligand. Using this newly developed fluorescence polarization-based assay to screen for candidate ligands, we identified the dietary molecules vanillin and ethylvanillin as weak bona fide MR1 ligands. Both upregulated MR1 on the surface of C1R.MR1 cells and the crystal structure of a MAIT cell T cell receptor-MR1-ethylvanillin complex revealed that ethylvanillin formed a Schiff base with K43 of MR1 and was buried within the A'-pocket. Collectively, we developed and validated a method to quantitate the binding affinities of ligands for MR1 that will enable an efficient and rapid screening of candidate MR1 ligands.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: